Manuel Wiesenfarth
Division of Biostatistics, German Cancer Research Center
Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge
Aug 10, 2023



Abstract:Quantitative organ assessment is an essential step in automated abdominal disease diagnosis and treatment planning. Artificial intelligence (AI) has shown great potential to automatize this process. However, most existing AI algorithms rely on many expert annotations and lack a comprehensive evaluation of accuracy and efficiency in real-world multinational settings. To overcome these limitations, we organized the FLARE 2022 Challenge, the largest abdominal organ analysis challenge to date, to benchmark fast, low-resource, accurate, annotation-efficient, and generalized AI algorithms. We constructed an intercontinental and multinational dataset from more than 50 medical groups, including Computed Tomography (CT) scans with different races, diseases, phases, and manufacturers. We independently validated that a set of AI algorithms achieved a median Dice Similarity Coefficient (DSC) of 90.0\% by using 50 labeled scans and 2000 unlabeled scans, which can significantly reduce annotation requirements. The best-performing algorithms successfully generalized to holdout external validation sets, achieving a median DSC of 89.5\%, 90.9\%, and 88.3\% on North American, European, and Asian cohorts, respectively. They also enabled automatic extraction of key organ biology features, which was labor-intensive with traditional manual measurements. This opens the potential to use unlabeled data to boost performance and alleviate annotation shortages for modern AI models.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022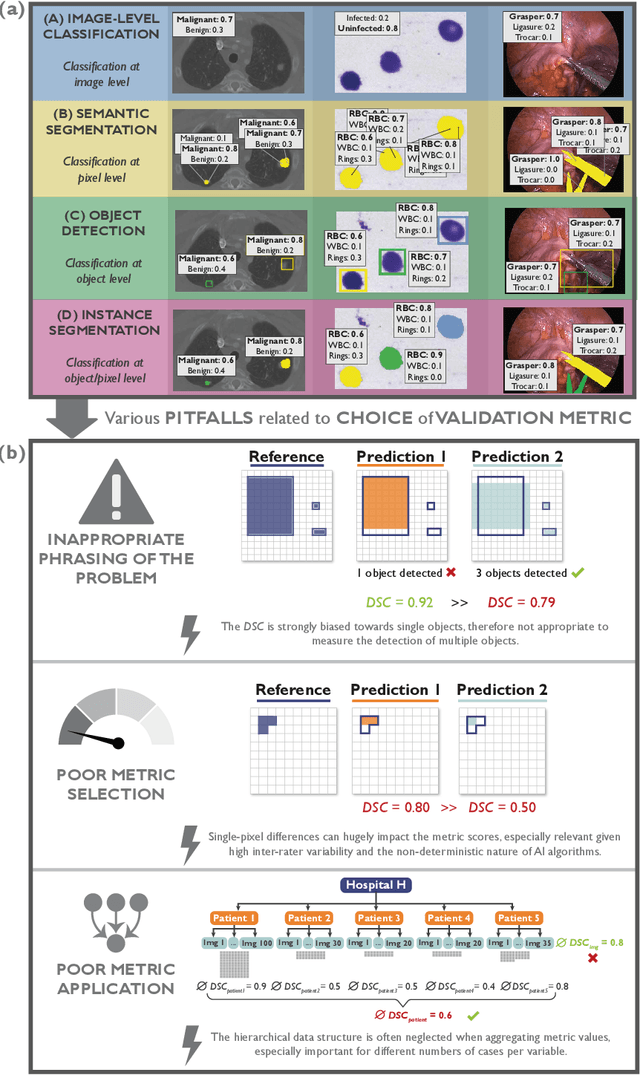
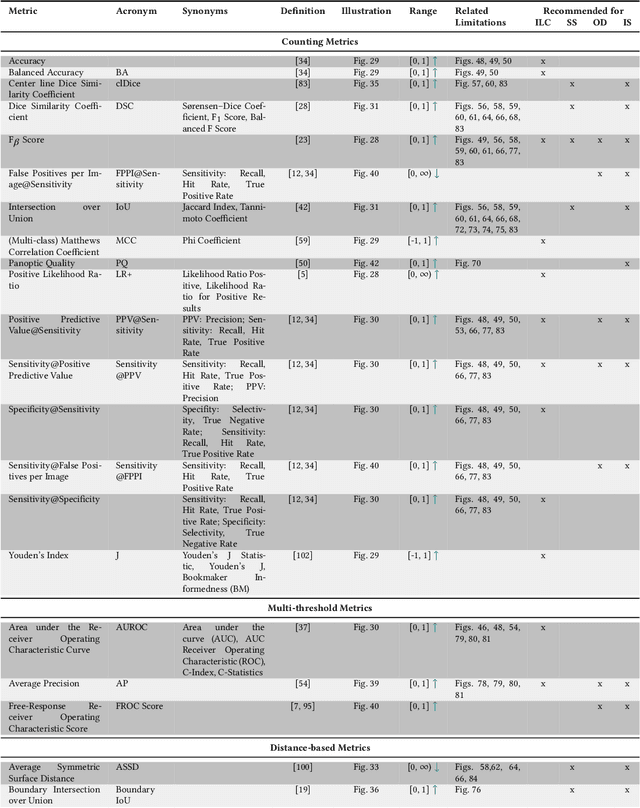
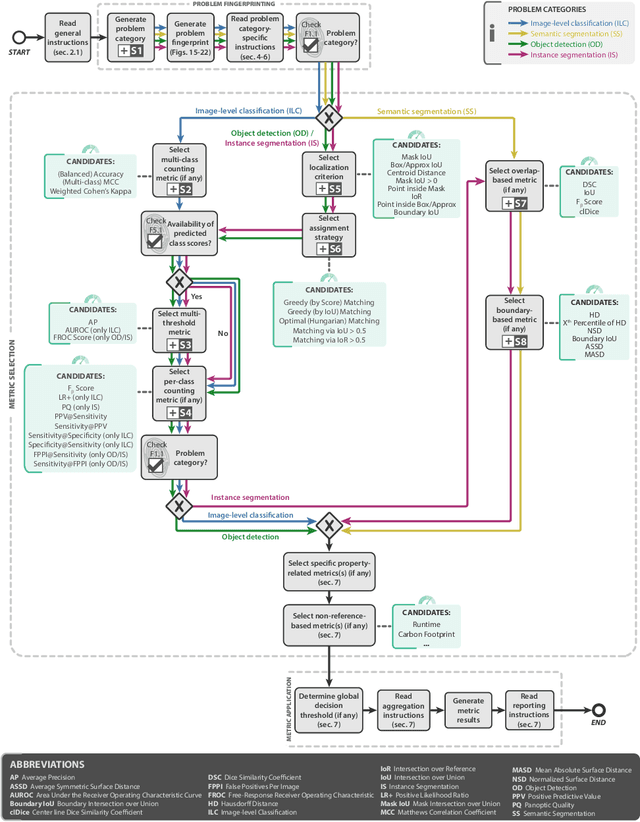
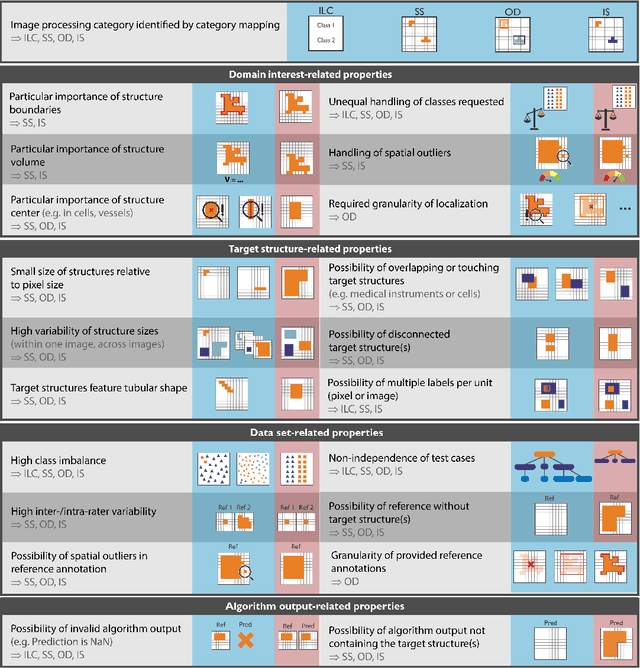
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
How can we learn from challenges? A statistical approach to driving future algorithm development
Jun 17, 2021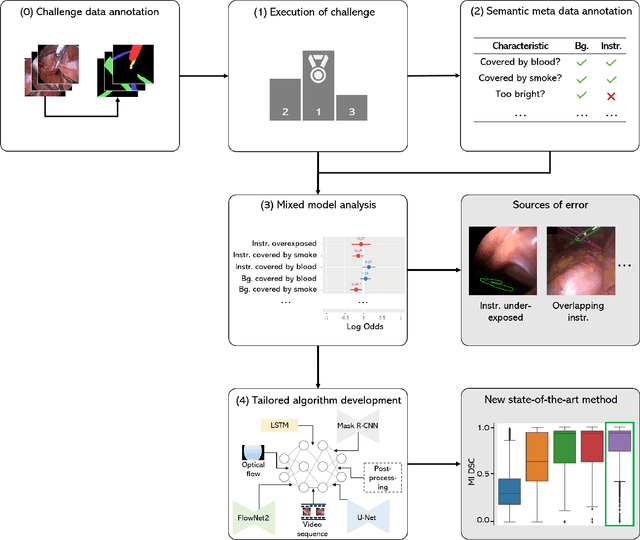
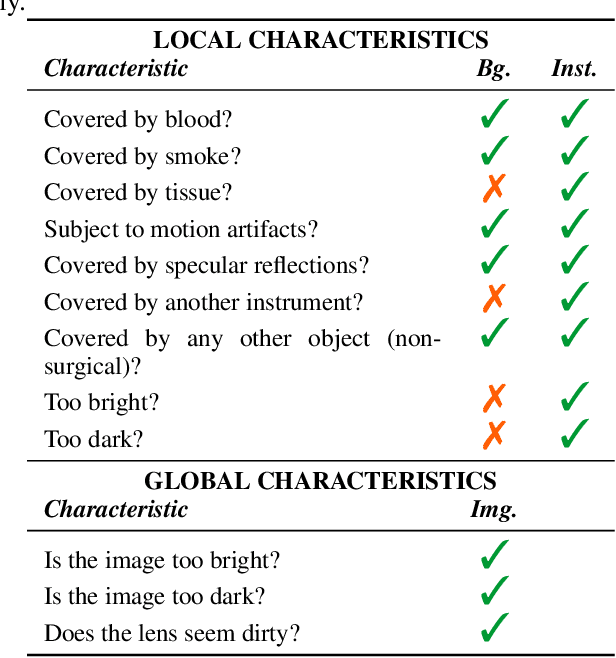
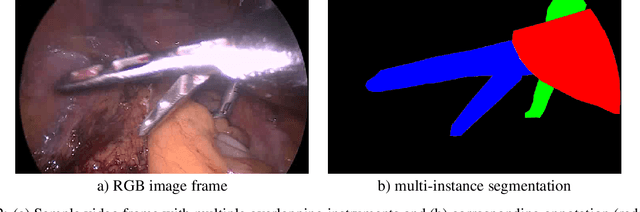
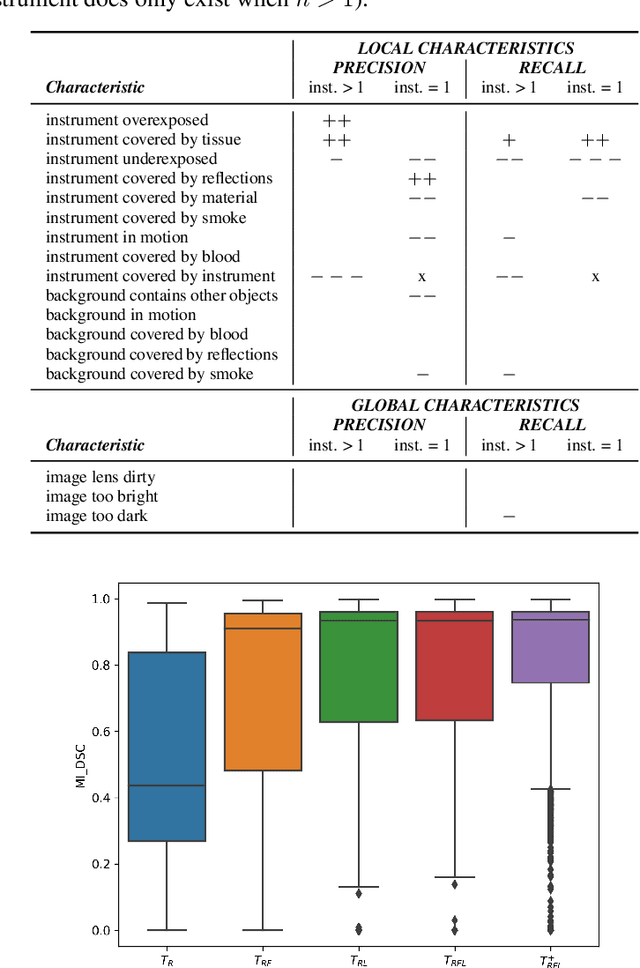
Abstract:Challenges have become the state-of-the-art approach to benchmark image analysis algorithms in a comparative manner. While the validation on identical data sets was a great step forward, results analysis is often restricted to pure ranking tables, leaving relevant questions unanswered. Specifically, little effort has been put into the systematic investigation on what characterizes images in which state-of-the-art algorithms fail. To address this gap in the literature, we (1) present a statistical framework for learning from challenges and (2) instantiate it for the specific task of instrument instance segmentation in laparoscopic videos. Our framework relies on the semantic meta data annotation of images, which serves as foundation for a General Linear Mixed Models (GLMM) analysis. Based on 51,542 meta data annotations performed on 2,728 images, we applied our approach to the results of the Robust Medical Instrument Segmentation Challenge (ROBUST-MIS) challenge 2019 and revealed underexposure, motion and occlusion of instruments as well as the presence of smoke or other objects in the background as major sources of algorithm failure. Our subsequent method development, tailored to the specific remaining issues, yielded a deep learning model with state-of-the-art overall performance and specific strengths in the processing of images in which previous methods tended to fail. Due to the objectivity and generic applicability of our approach, it could become a valuable tool for validation in the field of medical image analysis and beyond. and segmentation of small, crossing, moving and transparent instrument(s) (parts).
Machine learning-based analysis of hyperspectral images for automated sepsis diagnosis
Jun 15, 2021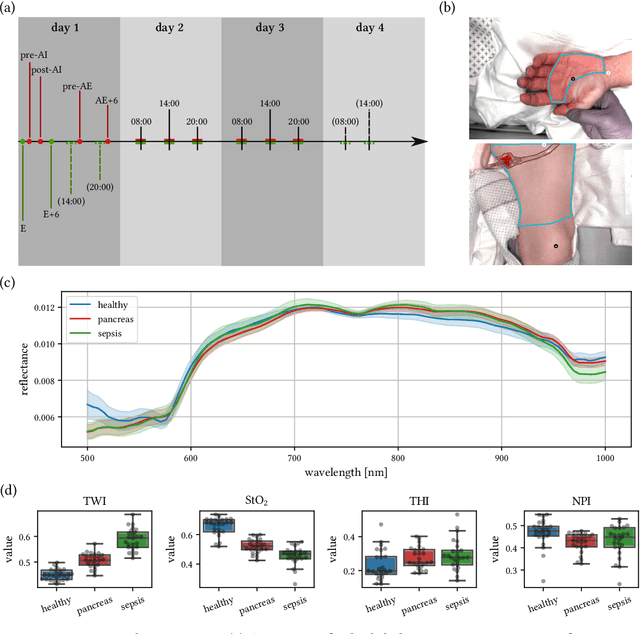
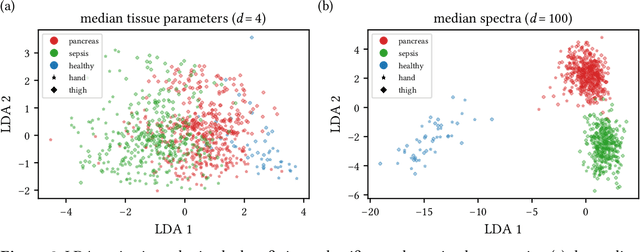
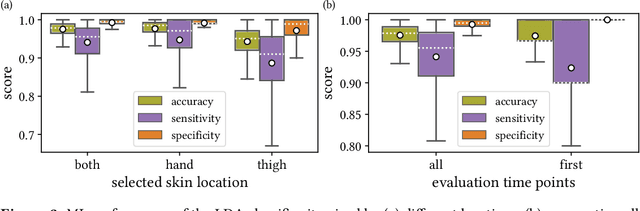

Abstract:Sepsis is a leading cause of mortality and critical illness worldwide. While robust biomarkers for early diagnosis are still missing, recent work indicates that hyperspectral imaging (HSI) has the potential to overcome this bottleneck by monitoring microcirculatory alterations. Automated machine learning-based diagnosis of sepsis based on HSI data, however, has not been explored to date. Given this gap in the literature, we leveraged an existing data set to (1) investigate whether HSI-based automated diagnosis of sepsis is possible and (2) put forth a list of possible confounders relevant for HSI-based tissue classification. While we were able to classify sepsis with an accuracy of over $98\,\%$ using the existing data, our research also revealed several subject-, therapy- and imaging-related confounders that may lead to an overestimation of algorithm performance when not balanced across the patient groups. We conclude that further prospective studies, carefully designed with respect to these confounders, are necessary to confirm the preliminary results obtained in this study.
The Medical Segmentation Decathlon
Jun 10, 2021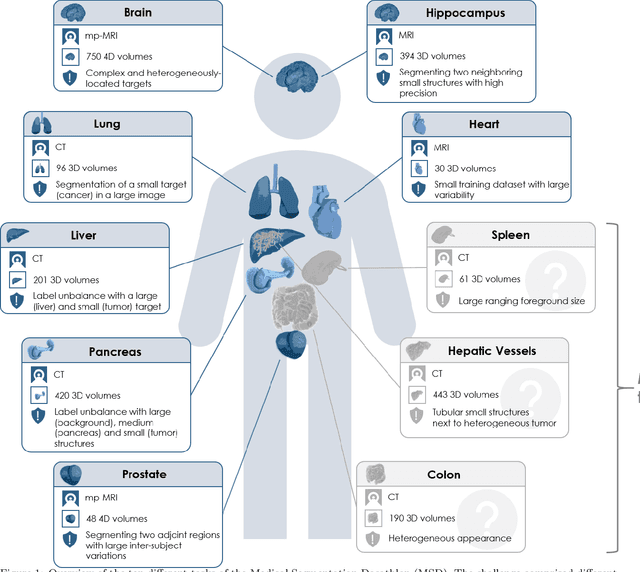
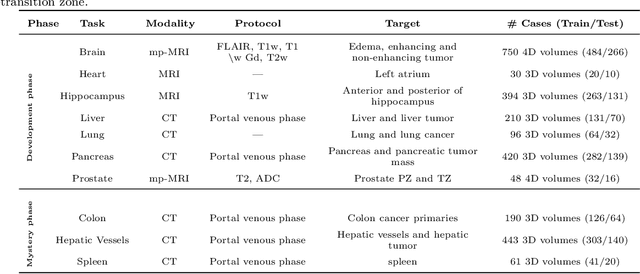
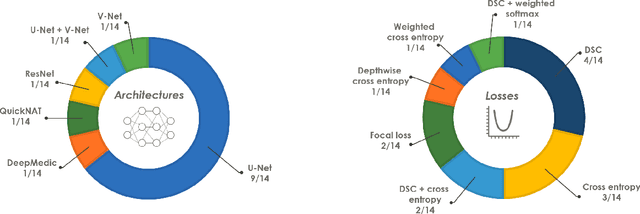
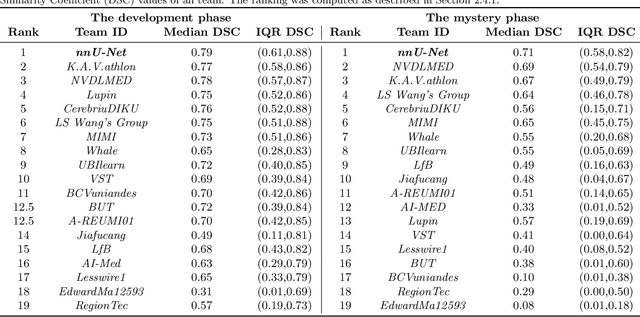
Abstract:International challenges have become the de facto standard for comparative assessment of image analysis algorithms given a specific task. Segmentation is so far the most widely investigated medical image processing task, but the various segmentation challenges have typically been organized in isolation, such that algorithm development was driven by the need to tackle a single specific clinical problem. We hypothesized that a method capable of performing well on multiple tasks will generalize well to a previously unseen task and potentially outperform a custom-designed solution. To investigate the hypothesis, we organized the Medical Segmentation Decathlon (MSD) - a biomedical image analysis challenge, in which algorithms compete in a multitude of both tasks and modalities. The underlying data set was designed to explore the axis of difficulties typically encountered when dealing with medical images, such as small data sets, unbalanced labels, multi-site data and small objects. The MSD challenge confirmed that algorithms with a consistent good performance on a set of tasks preserved their good average performance on a different set of previously unseen tasks. Moreover, by monitoring the MSD winner for two years, we found that this algorithm continued generalizing well to a wide range of other clinical problems, further confirming our hypothesis. Three main conclusions can be drawn from this study: (1) state-of-the-art image segmentation algorithms are mature, accurate, and generalize well when retrained on unseen tasks; (2) consistent algorithmic performance across multiple tasks is a strong surrogate of algorithmic generalizability; (3) the training of accurate AI segmentation models is now commoditized to non AI experts.
Robust Medical Instrument Segmentation Challenge 2019
Mar 23, 2020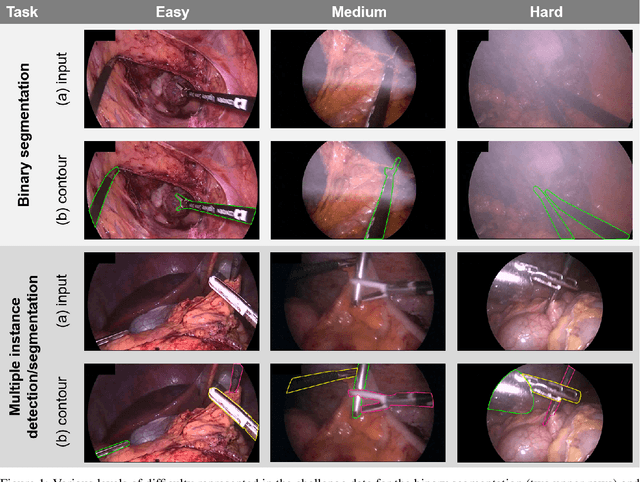
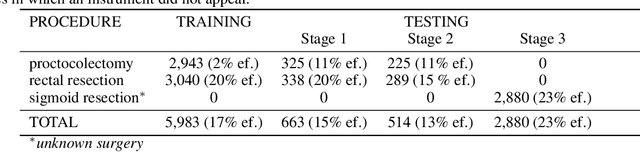
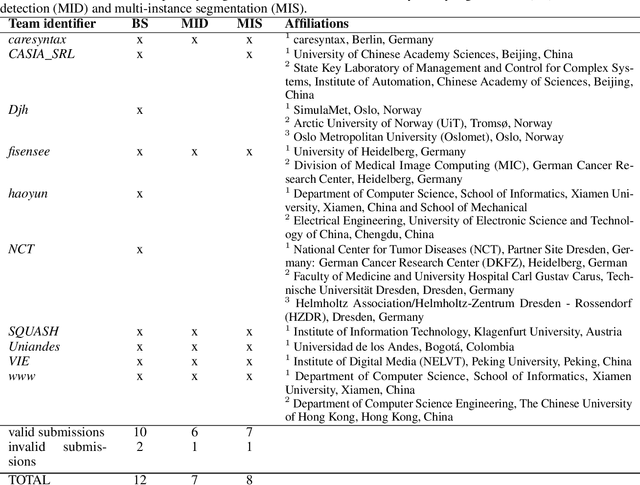
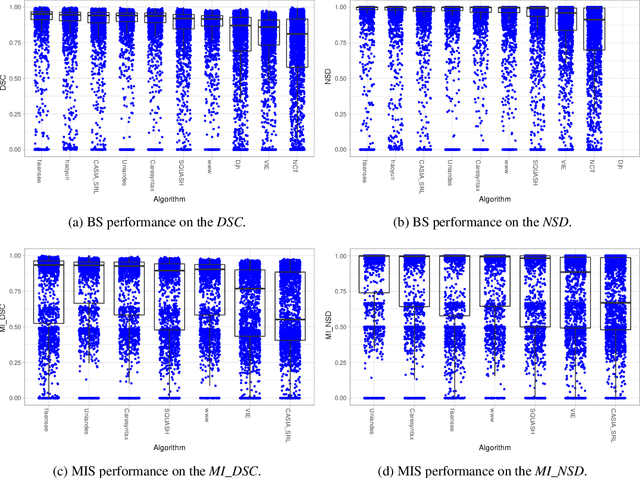
Abstract:Intraoperative tracking of laparoscopic instruments is often a prerequisite for computer and robotic-assisted interventions. While numerous methods for detecting, segmenting and tracking of medical instruments based on endoscopic video images have been proposed in the literature, key limitations remain to be addressed: Firstly, robustness, that is, the reliable performance of state-of-the-art methods when run on challenging images (e.g. in the presence of blood, smoke or motion artifacts). Secondly, generalization; algorithms trained for a specific intervention in a specific hospital should generalize to other interventions or institutions. In an effort to promote solutions for these limitations, we organized the Robust Medical Instrument Segmentation (ROBUST-MIS) challenge as an international benchmarking competition with a specific focus on the robustness and generalization capabilities of algorithms. For the first time in the field of endoscopic image processing, our challenge included a task on binary segmentation and also addressed multi-instance detection and segmentation. The challenge was based on a surgical data set comprising 10,040 annotated images acquired from a total of 30 surgical procedures from three different types of surgery. The validation of the competing methods for the three tasks (binary segmentation, multi-instance detection and multi-instance segmentation) was performed in three different stages with an increasing domain gap between the training and the test data. The results confirm the initial hypothesis, namely that algorithm performance degrades with an increasing domain gap. While the average detection and segmentation quality of the best-performing algorithms is high, future research should concentrate on detection and segmentation of small, crossing, moving and transparent instrument(s) (parts).
Methods and open-source toolkit for analyzing and visualizing challenge results
Oct 11, 2019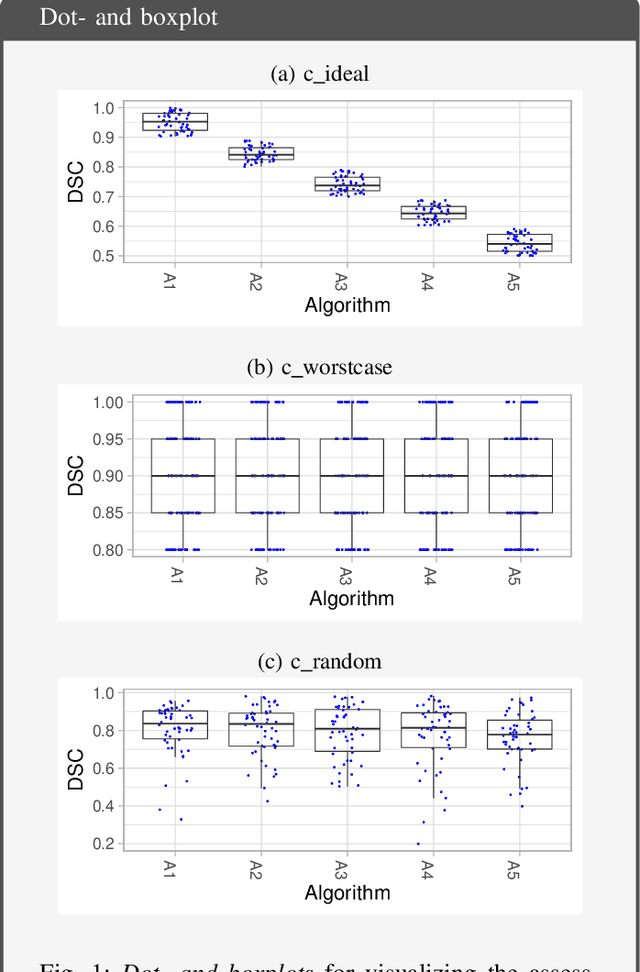

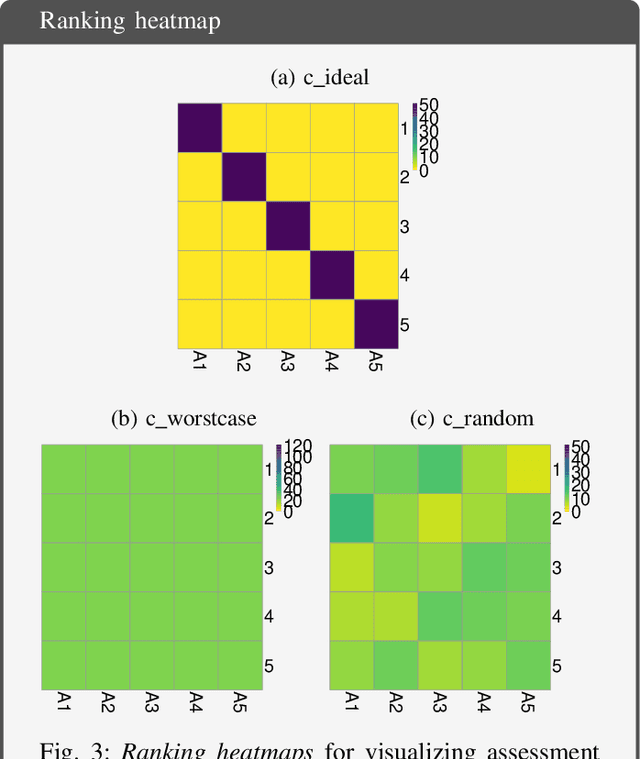
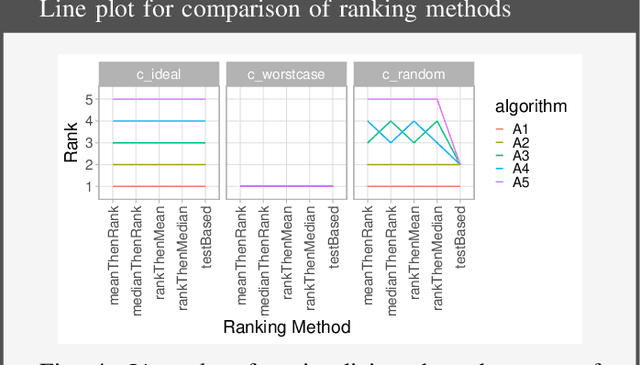
Abstract:Biomedical challenges have become the de facto standard for benchmarking biomedical image analysis algorithms. While the number of challenges is steadily increasing, surprisingly little effort has been invested in ensuring high quality design, execution and reporting for these international competitions. Specifically, results analysis and visualization in the event of uncertainties have been given almost no attention in the literature. Given these shortcomings, the contribution of this paper is two-fold: (1) We present a set of methods to comprehensively analyze and visualize the results of single-task and multi-task challenges and apply them to a number of simulated and real-life challenges to demonstrate their specific strengths and weaknesses; (2) We release the open-source framework challengeR as part of this work to enable fast and wide adoption of the methodology proposed in this paper. Our approach offers an intuitive way to gain important insights into the relative and absolute performance of algorithms, which cannot be revealed by commonly applied visualization techniques. This is demonstrated by the experiments performed within this work. Our framework could thus become an important tool for analyzing and visualizing challenge results in the field of biomedical image analysis and beyond.
Exploiting the potential of unlabeled endoscopic video data with self-supervised learning
Jan 31, 2018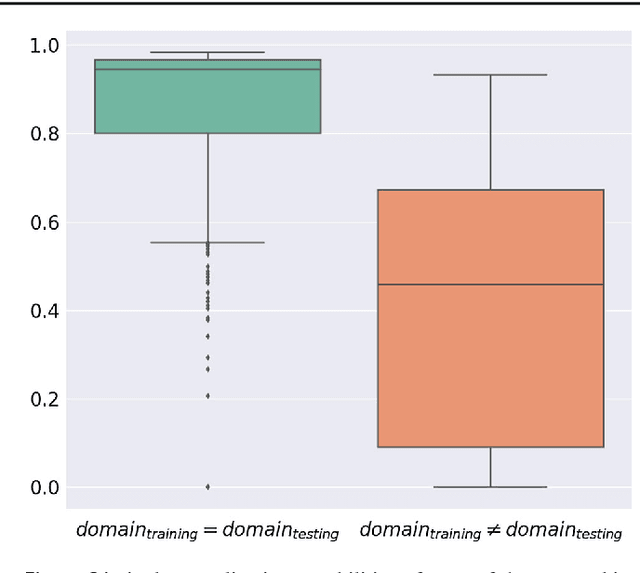
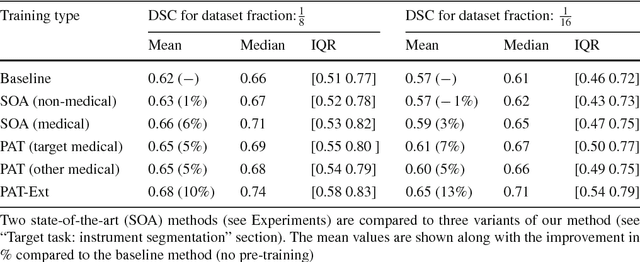
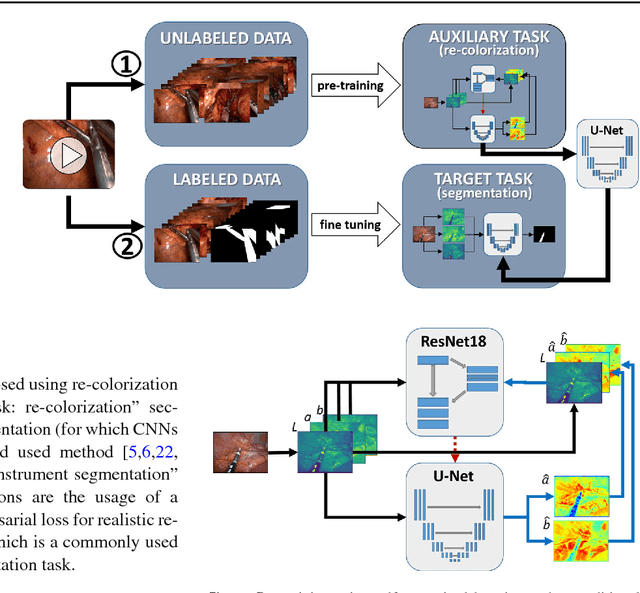
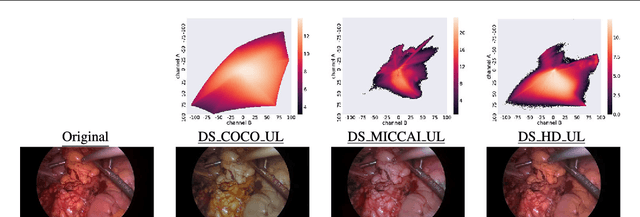
Abstract:Surgical data science is a new research field that aims to observe all aspects of the patient treatment process in order to provide the right assistance at the right time. Due to the breakthrough successes of deep learning-based solutions for automatic image annotation, the availability of reference annotations for algorithm training is becoming a major bottleneck in the field. The purpose of this paper was to investigate the concept of self-supervised learning to address this issue. Our approach is guided by the hypothesis that unlabeled video data can be used to learn a representation of the target domain that boosts the performance of state-of-the-art machine learning algorithms when used for pre-training. Core of the method is an auxiliary task based on raw endoscopic video data of the target domain that is used to initialize the convolutional neural network (CNN) for the target task. In this paper, we propose the re-colorization of medical images with a generative adversarial network (GAN)-based architecture as auxiliary task. A variant of the method involves a second pre-training step based on labeled data for the target task from a related domain. We validate both variants using medical instrument segmentation as target task. The proposed approach can be used to radically reduce the manual annotation effort involved in training CNNs. Compared to the baseline approach of generating annotated data from scratch, our method decreases exploratively the number of labeled images by up to 75% without sacrificing performance. Our method also outperforms alternative methods for CNN pre-training, such as pre-training on publicly available non-medical or medical data using the target task (in this instance: segmentation). As it makes efficient use of available (non-)public and (un-)labeled data, the approach has the potential to become a valuable tool for CNN (pre-)training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge