Minu D. Tizabi
False Promises in Medical Imaging AI? Assessing Validity of Outperformance Claims
May 07, 2025Abstract:Performance comparisons are fundamental in medical imaging Artificial Intelligence (AI) research, often driving claims of superiority based on relative improvements in common performance metrics. However, such claims frequently rely solely on empirical mean performance. In this paper, we investigate whether newly proposed methods genuinely outperform the state of the art by analyzing a representative cohort of medical imaging papers. We quantify the probability of false claims based on a Bayesian approach that leverages reported results alongside empirically estimated model congruence to estimate whether the relative ranking of methods is likely to have occurred by chance. According to our results, the majority (>80%) of papers claims outperformance when introducing a new method. Our analysis further revealed a high probability (>5%) of false outperformance claims in 86% of classification papers and 53% of segmentation papers. These findings highlight a critical flaw in current benchmarking practices: claims of outperformance in medical imaging AI are frequently unsubstantiated, posing a risk of misdirecting future research efforts.
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024



Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
Quality Assured: Rethinking Annotation Strategies in Imaging AI
Jul 26, 2024



Abstract:This paper does not describe a novel method. Instead, it studies an essential foundation for reliable benchmarking and ultimately real-world application of AI-based image analysis: generating high-quality reference annotations. Previous research has focused on crowdsourcing as a means of outsourcing annotations. However, little attention has so far been given to annotation companies, specifically regarding their internal quality assurance (QA) processes. Therefore, our aim is to evaluate the influence of QA employed by annotation companies on annotation quality and devise methodologies for maximizing data annotation efficacy. Based on a total of 57,648 instance segmented images obtained from a total of 924 annotators and 34 QA workers from four annotation companies and Amazon Mechanical Turk (MTurk), we derived the following insights: (1) Annotation companies perform better both in terms of quantity and quality compared to the widely used platform MTurk. (2) Annotation companies' internal QA only provides marginal improvements, if any. However, improving labeling instructions instead of investing in QA can substantially boost annotation performance. (3) The benefit of internal QA depends on specific image characteristics. Our work could enable researchers to derive substantially more value from a fixed annotation budget and change the way annotation companies conduct internal QA.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Labeling instructions matter in biomedical image analysis
Jul 20, 2022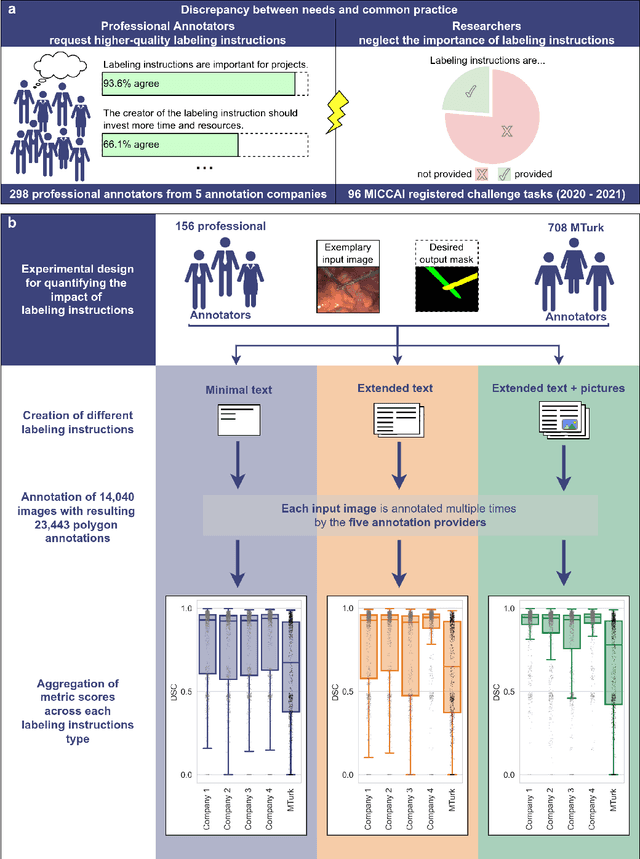
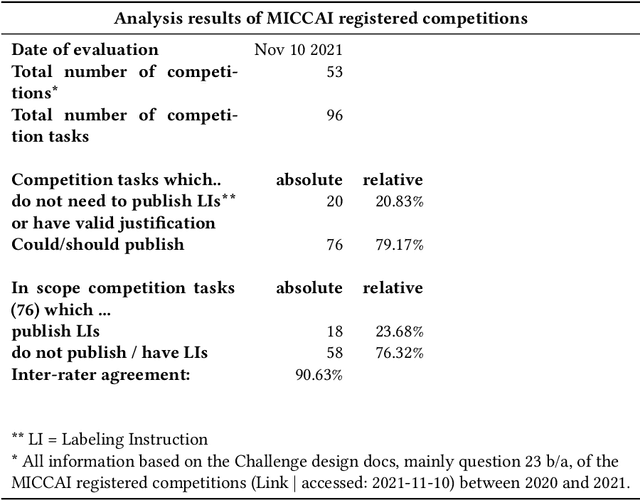
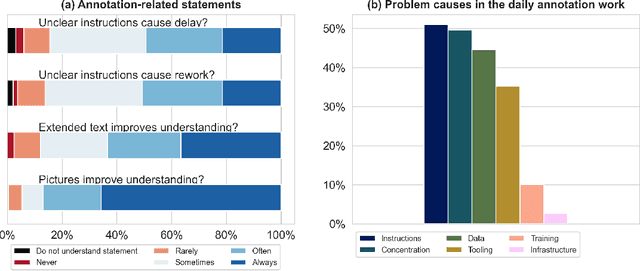
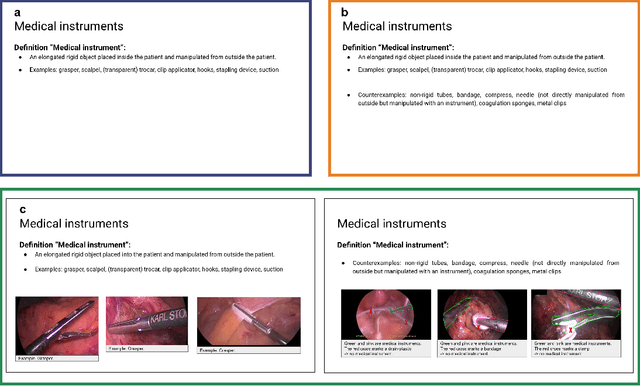
Abstract:Biomedical image analysis algorithm validation depends on high-quality annotation of reference datasets, for which labeling instructions are key. Despite their importance, their optimization remains largely unexplored. Here, we present the first systematic study of labeling instructions and their impact on annotation quality in the field. Through comprehensive examination of professional practice and international competitions registered at the MICCAI Society, we uncovered a discrepancy between annotators' needs for labeling instructions and their current quality and availability. Based on an analysis of 14,040 images annotated by 156 annotators from four professional companies and 708 Amazon Mechanical Turk (MTurk) crowdworkers using instructions with different information density levels, we further found that including exemplary images significantly boosts annotation performance compared to text-only descriptions, while solely extending text descriptions does not. Finally, professional annotators constantly outperform MTurk crowdworkers. Our study raises awareness for the need of quality standards in biomedical image analysis labeling instructions.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022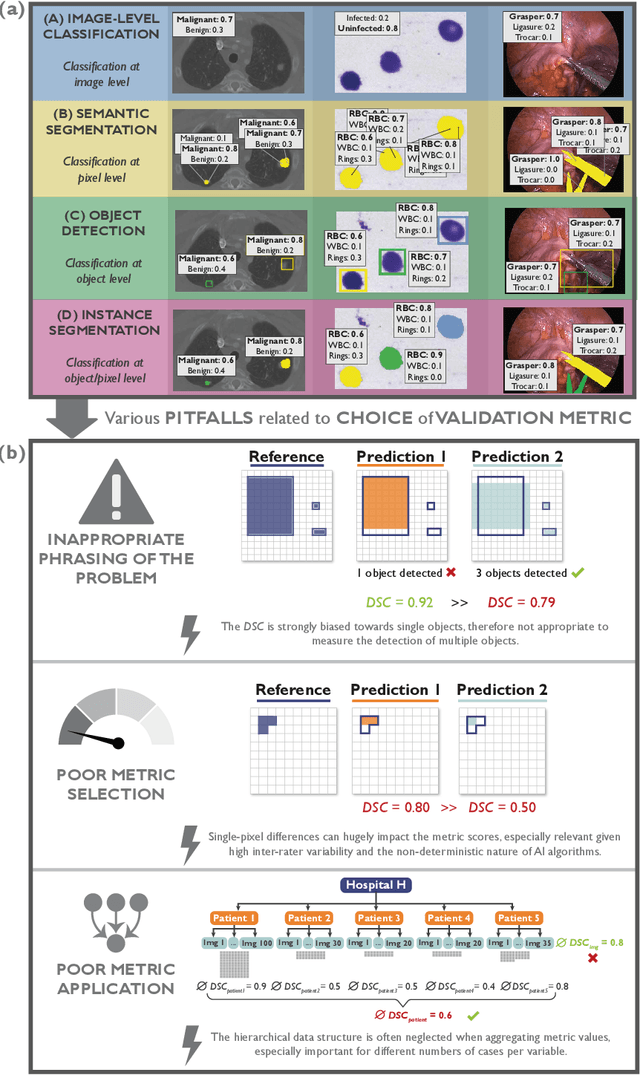
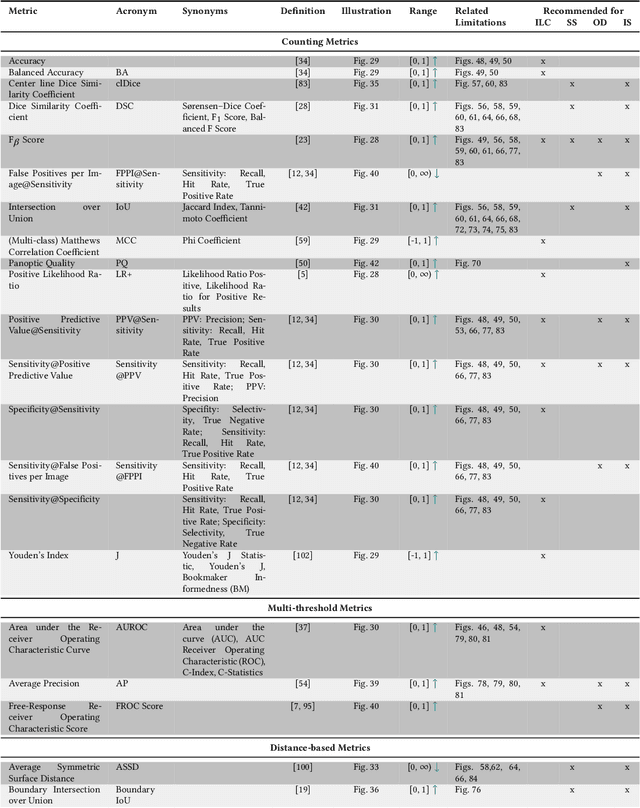
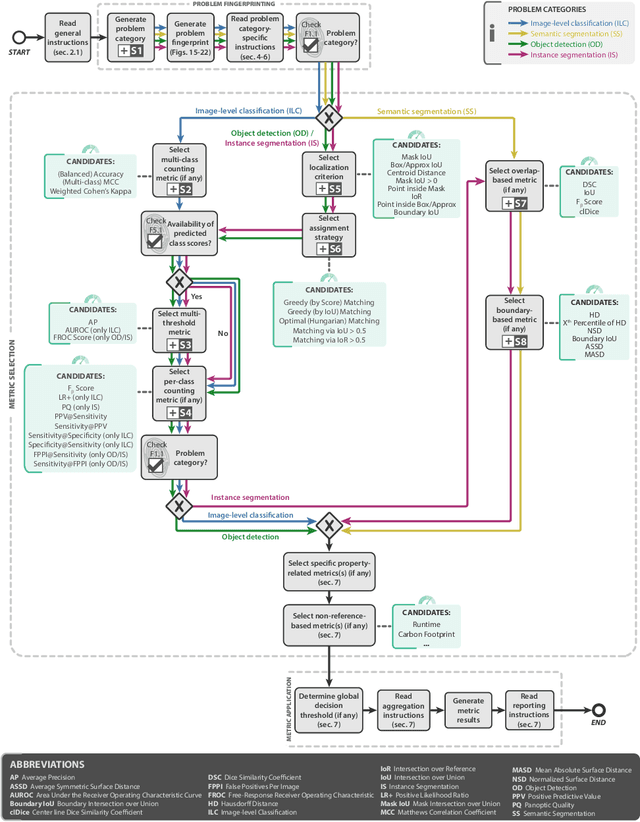
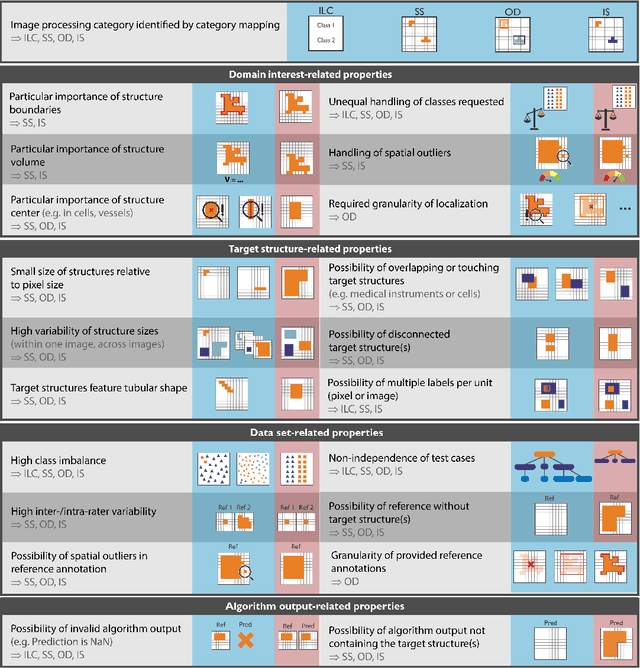
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
Semantic segmentation of multispectral photoacoustic images using deep learning
May 20, 2021
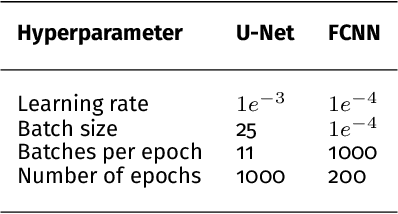

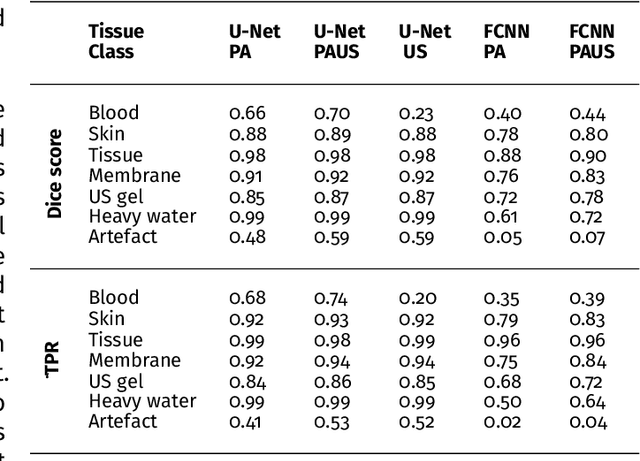
Abstract:Photoacoustic imaging has the potential to revolutionise healthcare due to the valuable information on tissue physiology that is contained in multispectral photoacoustic measurements. Clinical translation of the technology requires conversion of the high-dimensional acquired data into clinically relevant and interpretable information. In this work, we present a deep learning-based approach to semantic segmentation of multispectral photoacoustic images to facilitate the interpretability of recorded images. Manually annotated multispectral photoacoustic imaging data are used as gold standard reference annotations and enable the training of a deep learning-based segmentation algorithm in a supervised manner. Based on a validation study with experimentally acquired data of healthy human volunteers, we show that automatic tissue segmentation can be used to create powerful analyses and visualisations of multispectral photoacoustic images. Due to the intuitive representation of high-dimensional information, such a processing algorithm could be a valuable means to facilitate the clinical translation of photoacoustic imaging.
Common Limitations of Image Processing Metrics: A Picture Story
Apr 13, 2021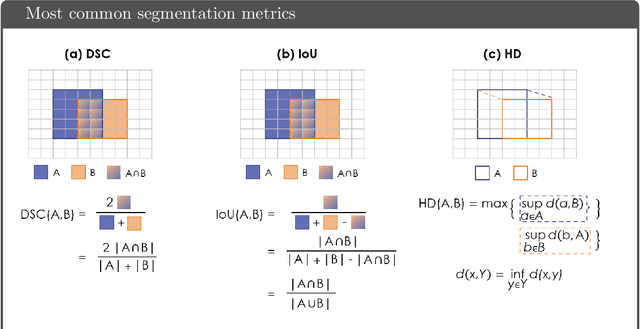
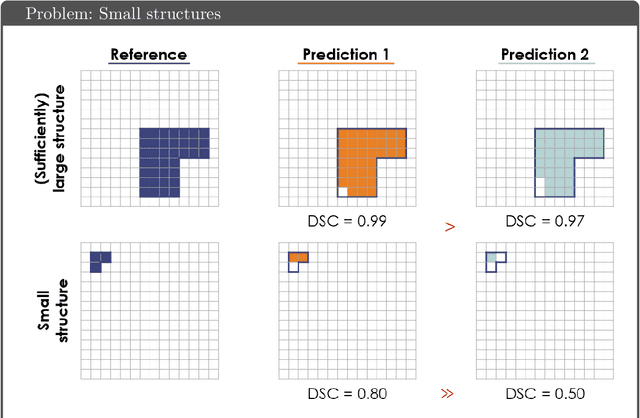
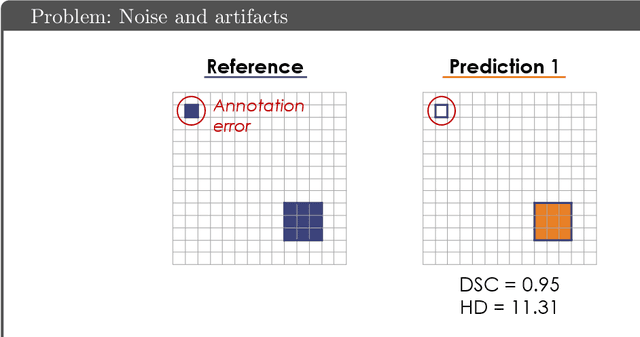
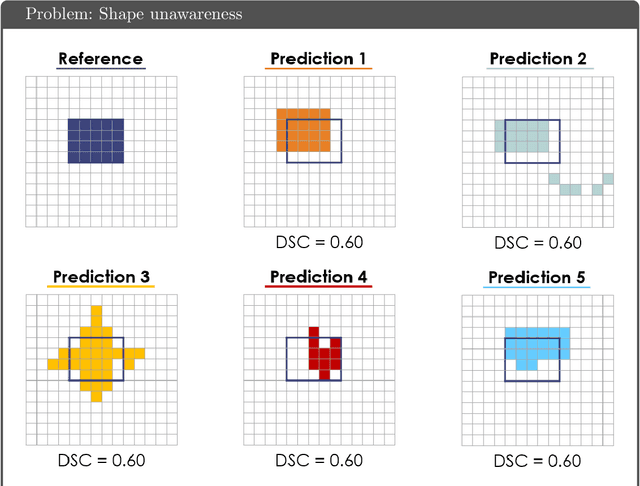
Abstract:While the importance of automatic image analysis is increasing at an enormous pace, recent meta-research revealed major flaws with respect to algorithm validation. Specifically, performance metrics are key for objective, transparent and comparative performance assessment, but relatively little attention has been given to the practical pitfalls when using specific metrics for a given image analysis task. A common mission of several international initiatives is therefore to provide researchers with guidelines and tools to choose the performance metrics in a problem-aware manner. This dynamically updated document has the purpose to illustrate important limitations of performance metrics commonly applied in the field of image analysis. The current version is based on a Delphi process on metrics conducted by an international consortium of image analysis experts.
Data-driven generation of plausible tissue geometries for realistic photoacoustic image synthesis
Mar 29, 2021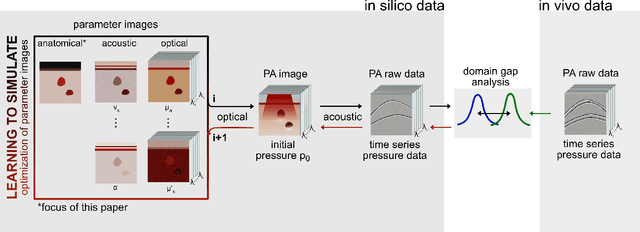
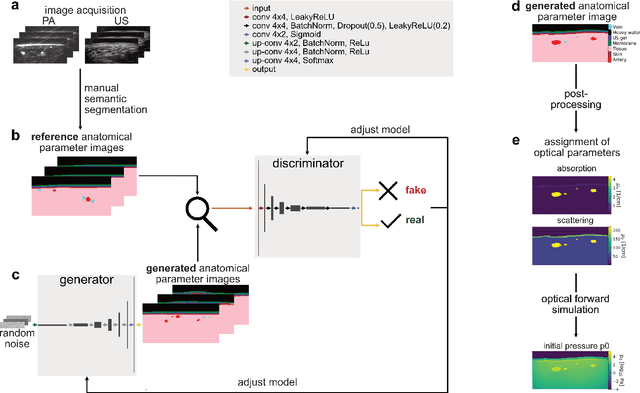
Abstract:Photoacoustic tomography (PAT) has the potential to recover morphological and functional tissue properties such as blood oxygenation with high spatial resolution and in an interventional setting. However, decades of research invested in solving the inverse problem of recovering clinically relevant tissue properties from spectral measurements have failed to produce solutions that can quantify tissue parameters robustly in a clinical setting. Previous attempts to address the limitations of model-based approaches with machine learning were hampered by the absence of labeled reference data needed for supervised algorithm training. While this bottleneck has been tackled by simulating training data, the domain gap between real and simulated images remains a huge unsolved challenge. As a first step to address this bottleneck, we propose a novel approach to PAT data simulation, which we refer to as "learning to simulate". Our approach involves subdividing the challenge of generating plausible simulations into two disjoint problems: (1) Probabilistic generation of realistic tissue morphology, represented by semantic segmentation maps and (2) pixel-wise assignment of corresponding optical and acoustic properties. In the present work, we focus on the first challenge. Specifically, we leverage the concept of Generative Adversarial Networks (GANs) trained on semantically annotated medical imaging data to generate plausible tissue geometries. According to an initial in silico feasibility study our approach is well-suited for contributing to realistic PAT image synthesis and could thus become a fundamental step for deep learning-based quantitative PAT.
Tattoo tomography: Freehand 3D photoacoustic image reconstruction with an optical pattern
Nov 11, 2020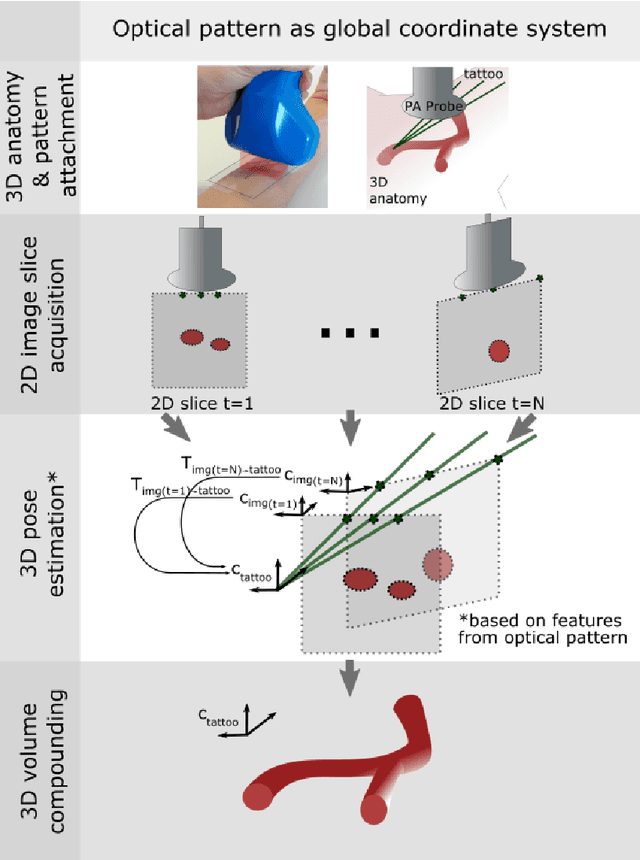
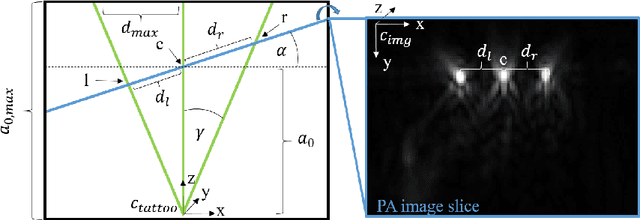
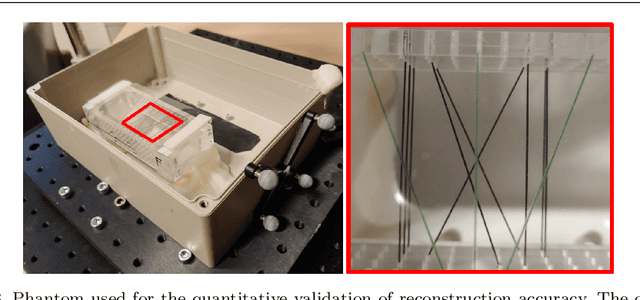

Abstract:Purpose: Photoacoustic tomography (PAT) is a novel imaging technique that can spatially resolve both morphological and functional tissue properties, such as the vessel topology and tissue oxygenation. While this capacity makes PAT a promising modality for the diagnosis, treatment and follow-up of various diseases, a current drawback is the limited field-of-view (FoV) provided by the conventionally applied 2D probes. Methods: In this paper, we present a novel approach to 3D reconstruction of PAT data (Tattoo tomography) that does not require an external tracking system and can smoothly be integrated into clinical workflows. It is based on an optical pattern placed on the region of interest prior to image acquisition. This pattern is designed in a way that a tomographic image of it enables the recovery of the probe pose relative to the coordinate system of the pattern. This allows the transformation of a sequence of acquired PA images into one common global coordinate system and thus the consistent 3D reconstruction of PAT imaging data. Results: An initial feasibility study conducted with experimental phantom data and in vivo forearm data indicates that the Tattoo approach is well-suited for 3D reconstruction of PAT data with high accuracy and precision. Conclusion: In contrast to previous approaches to 3D ultrasound (US) or PAT reconstruction, the Tattoo approach neither requires complex external hardware nor training data acquired for a specific application. It could thus become a valuable tool for clinical freehand PAT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge