Bjoern Menze
for the ALFA study
SigVLP: Sigmoid Volume-Language Pre-Training for Self-Supervised CT-Volume Adaptive Representation Learning
Feb 25, 2026Abstract:Large-scale, volumetric medical imaging datasets typically aggregate scans from different vendors and devices, resulting in highly variable resolution, slice thicknesses, and numbers of slices per study. Consequently, training representation models usually requires cropping or interpolating along the z-axis to obtain fixed-size blocks, which inevitably causes information loss. We propose a new training approach to overcome this limitation. Instead of absolute position embeddings, we interpret volumes as sequences of 3D chunks and adopt Rotary Position Embeddings, allowing us to treat the z-axis as an unconstrained temporal dimensions. Building on this idea, we introduce a new vision-language model: SigVLP. In SigVLP, we implement Rotary Position Embedding as the positional encoding method, which is applied directly within the attention operation, generating input-conditioned sine and cosine weights on the fly. This design ensures consistent alignment between query and key projections and adapts to any input sizes. To allow for variable input size during training, we sample Computed Tomography volumes in chunks and pair them with localized organ-wise textual observations. Compared to using entire reports for conditioning, chunkwise alignment provides finer-grained supervision, enabling the model to establish stronger correlations between the text and volume representations, thereby improving the precision of text-to-volume alignment. Our models are trained with the Muon optimizer and evaluated on a diverse set of downstream tasks, including zero-shot abnormality and organ classification, segmentation, and retrieval tasks.
VariViT: A Vision Transformer for Variable Image Sizes
Feb 16, 2026Abstract:Vision Transformers (ViTs) have emerged as the state-of-the-art architecture in representation learning, leveraging self-attention mechanisms to excel in various tasks. ViTs split images into fixed-size patches, constraining them to a predefined size and necessitating pre-processing steps like resizing, padding, or cropping. This poses challenges in medical imaging, particularly with irregularly shaped structures like tumors. A fixed bounding box crop size produces input images with highly variable foreground-to-background ratios. Resizing medical images can degrade information and introduce artefacts, impacting diagnosis. Hence, tailoring variable-sized crops to regions of interest can enhance feature representation capabilities. Moreover, large images are computationally expensive, and smaller sizes risk information loss, presenting a computation-accuracy tradeoff. We propose VariViT, an improved ViT model crafted to handle variable image sizes while maintaining a consistent patch size. VariViT employs a novel positional embedding resizing scheme for a variable number of patches. We also implement a new batching strategy within VariViT to reduce computational complexity, resulting in faster training and inference times. In our evaluations on two 3D brain MRI datasets, VariViT surpasses vanilla ViTs and ResNet in glioma genotype prediction and brain tumor classification. It achieves F1-scores of 75.5% and 76.3%, respectively, learning more discriminative features. Our proposed batching strategy reduces computation time by up to 30% compared to conventional architectures. These findings underscore the efficacy of VariViT in image representation learning. Our code can be found here: https://github.com/Aswathi-Varma/varivit
ProGiDiff: Prompt-Guided Diffusion-Based Medical Image Segmentation
Jan 22, 2026Abstract:Widely adopted medical image segmentation methods, although efficient, are primarily deterministic and remain poorly amenable to natural language prompts. Thus, they lack the capability to estimate multiple proposals, human interaction, and cross-modality adaptation. Recently, text-to-image diffusion models have shown potential to bridge the gap. However, training them from scratch requires a large dataset-a limitation for medical image segmentation. Furthermore, they are often limited to binary segmentation and cannot be conditioned on a natural language prompt. To this end, we propose a novel framework called ProGiDiff that leverages existing image generation models for medical image segmentation purposes. Specifically, we propose a ControlNet-style conditioning mechanism with a custom encoder, suitable for image conditioning, to steer a pre-trained diffusion model to output segmentation masks. It naturally extends to a multi-class setting simply by prompting the target organ. Our experiment on organ segmentation from CT images demonstrates strong performance compared to previous methods and could greatly benefit from an expert-in-the-loop setting to leverage multiple proposals. Importantly, we demonstrate that the learned conditioning mechanism can be easily transferred through low-rank, few-shot adaptation to segment MR images.
VERIDAH: Solving Enumeration Anomaly Aware Vertebra Labeling across Imaging Sequences
Jan 20, 2026Abstract:The human spine commonly consists of seven cervical, twelve thoracic, and five lumbar vertebrae. However, enumeration anomalies may result in individuals having eleven or thirteen thoracic vertebrae and four or six lumbar vertebrae. Although the identification of enumeration anomalies has potential clinical implications for chronic back pain and operation planning, the thoracolumbar junction is often poorly assessed and rarely described in clinical reports. Additionally, even though multiple deep-learning-based vertebra labeling algorithms exist, there is a lack of methods to automatically label enumeration anomalies. Our work closes that gap by introducing "Vertebra Identification with Anomaly Handling" (VERIDAH), a novel vertebra labeling algorithm based on multiple classification heads combined with a weighted vertebra sequence prediction algorithm. We show that our approach surpasses existing models on T2w TSE sagittal (98.30% vs. 94.24% of subjects with all vertebrae correctly labeled, p < 0.001) and CT imaging (99.18% vs. 77.26% of subjects with all vertebrae correctly labeled, p < 0.001) and works in arbitrary field-of-view images. VERIDAH correctly labeled the presence 2 Möller et al. of thoracic enumeration anomalies in 87.80% and 96.30% of T2w and CT images, respectively, and lumbar enumeration anomalies in 94.48% and 97.22% for T2w and CT, respectively. Our code and models are available at: https://github.com/Hendrik-code/spineps.
In search of truth: Evaluating concordance of AI-based anatomy segmentation models
Dec 17, 2025



Abstract:Purpose AI-based methods for anatomy segmentation can help automate characterization of large imaging datasets. The growing number of similar in functionality models raises the challenge of evaluating them on datasets that do not contain ground truth annotations. We introduce a practical framework to assist in this task. Approach We harmonize the segmentation results into a standard, interoperable representation, which enables consistent, terminology-based labeling of the structures. We extend 3D Slicer to streamline loading and comparison of these harmonized segmentations, and demonstrate how standard representation simplifies review of the results using interactive summary plots and browser-based visualization using OHIF Viewer. To demonstrate the utility of the approach we apply it to evaluating segmentation of 31 anatomical structures (lungs, vertebrae, ribs, and heart) by six open-source models - TotalSegmentator 1.5 and 2.6, Auto3DSeg, MOOSE, MultiTalent, and CADS - for a sample of Computed Tomography (CT) scans from the publicly available National Lung Screening Trial (NLST) dataset. Results We demonstrate the utility of the framework in enabling automating loading, structure-wise inspection and comparison across models. Preliminary results ascertain practical utility of the approach in allowing quick detection and review of problematic results. The comparison shows excellent agreement segmenting some (e.g., lung) but not all structures (e.g., some models produce invalid vertebrae or rib segmentations). Conclusions The resources developed are linked from https://imagingdatacommons.github.io/segmentation-comparison/ including segmentation harmonization scripts, summary plots, and visualization tools. This work assists in model evaluation in absence of ground truth, ultimately enabling informed model selection.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
Better Tokens for Better 3D: Advancing Vision-Language Modeling in 3D Medical Imaging
Oct 23, 2025Abstract:Recent progress in vision-language modeling for 3D medical imaging has been fueled by large-scale computed tomography (CT) corpora with paired free-text reports, stronger architectures, and powerful pretrained models. This has enabled applications such as automated report generation and text-conditioned 3D image synthesis. Yet, current approaches struggle with high-resolution, long-sequence volumes: contrastive pretraining often yields vision encoders that are misaligned with clinical language, and slice-wise tokenization blurs fine anatomy, reducing diagnostic performance on downstream tasks. We introduce BTB3D (Better Tokens for Better 3D), a causal convolutional encoder-decoder that unifies 2D and 3D training and inference while producing compact, frequency-aware volumetric tokens. A three-stage training curriculum enables (i) local reconstruction, (ii) overlapping-window tiling, and (iii) long-context decoder refinement, during which the model learns from short slice excerpts yet generalizes to scans exceeding 300 slices without additional memory overhead. BTB3D sets a new state-of-the-art on two key tasks: it improves BLEU scores and increases clinical F1 by 40% over CT2Rep, CT-CHAT, and Merlin for report generation; and it reduces FID by 75% and halves FVD compared to GenerateCT and MedSyn for text-to-CT synthesis, producing anatomically consistent 512*512*241 volumes. These results confirm that precise three-dimensional tokenization, rather than larger language backbones alone, is essential for scalable vision-language modeling in 3D medical imaging. The codebase is available at: https://github.com/ibrahimethemhamamci/BTB3D
fastWDM3D: Fast and Accurate 3D Healthy Tissue Inpainting
Jul 17, 2025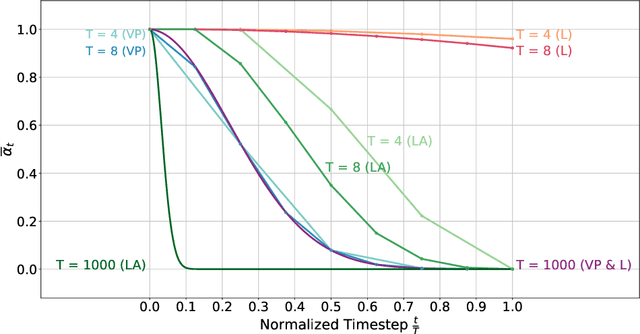
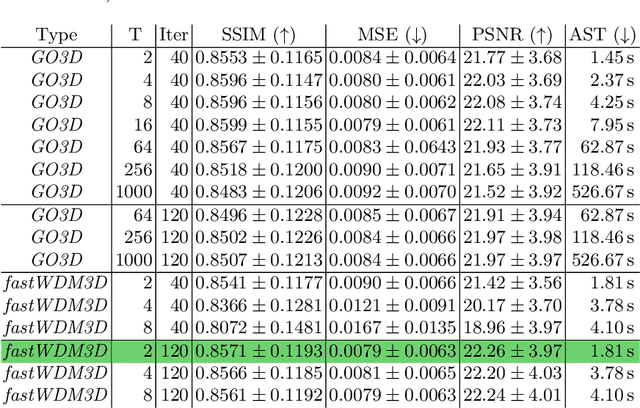
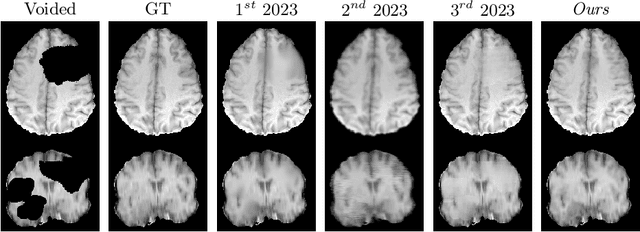
Abstract:Healthy tissue inpainting has significant applications, including the generation of pseudo-healthy baselines for tumor growth models and the facilitation of image registration. In previous editions of the BraTS Local Synthesis of Healthy Brain Tissue via Inpainting Challenge, denoising diffusion probabilistic models (DDPMs) demonstrated qualitatively convincing results but suffered from low sampling speed. To mitigate this limitation, we adapted a 2D image generation approach, combining DDPMs with generative adversarial networks (GANs) and employing a variance-preserving noise schedule, for the task of 3D inpainting. Our experiments showed that the variance-preserving noise schedule and the selected reconstruction losses can be effectively utilized for high-quality 3D inpainting in a few time steps without requiring adversarial training. We applied our findings to a different architecture, a 3D wavelet diffusion model (WDM3D) that does not include a GAN component. The resulting model, denoted as fastWDM3D, obtained a SSIM of 0.8571, a MSE of 0.0079, and a PSNR of 22.26 on the BraTS inpainting test set. Remarkably, it achieved these scores using only two time steps, completing the 3D inpainting process in 1.81 s per image. When compared to other DDPMs used for healthy brain tissue inpainting, our model is up to 800 x faster while still achieving superior performance metrics. Our proposed method, fastWDM3D, represents a promising approach for fast and accurate healthy tissue inpainting. Our code is available at https://github.com/AliciaDurrer/fastWDM3D.
Parametric shape models for vessels learned from segmentations via differentiable voxelization
Jul 03, 2025Abstract:Vessels are complex structures in the body that have been studied extensively in multiple representations. While voxelization is the most common of them, meshes and parametric models are critical in various applications due to their desirable properties. However, these representations are typically extracted through segmentations and used disjointly from each other. We propose a framework that joins the three representations under differentiable transformations. By leveraging differentiable voxelization, we automatically extract a parametric shape model of the vessels through shape-to-segmentation fitting, where we learn shape parameters from segmentations without the explicit need for ground-truth shape parameters. The vessel is parametrized as centerlines and radii using cubic B-splines, ensuring smoothness and continuity by construction. Meshes are differentiably extracted from the learned shape parameters, resulting in high-fidelity meshes that can be manipulated post-fit. Our method can accurately capture the geometry of complex vessels, as demonstrated by the volumetric fits in experiments on aortas, aneurysms, and brain vessels.
CRG Score: A Distribution-Aware Clinical Metric for Radiology Report Generation
May 22, 2025Abstract:Evaluating long-context radiology report generation is challenging. NLG metrics fail to capture clinical correctness, while LLM-based metrics often lack generalizability. Clinical accuracy metrics are more relevant but are sensitive to class imbalance, frequently favoring trivial predictions. We propose the CRG Score, a distribution-aware and adaptable metric that evaluates only clinically relevant abnormalities explicitly described in reference reports. CRG supports both binary and structured labels (e.g., type, location) and can be paired with any LLM for feature extraction. By balancing penalties based on label distribution, it enables fairer, more robust evaluation and serves as a clinically aligned reward function.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge