Jiacheng Wang
Sherman
Wireless Context Engineering for Efficient Mobile Agentic AI and Edge General Intelligence
Feb 07, 2026Abstract:Future wireless networks demand increasingly powerful intelligence to support sensing, communication, and autonomous decision-making. While scaling laws suggest improving performance by enlarging model capacity, practical edge deployments are fundamentally constrained by latency, energy, and memory, making unlimited model scaling infeasible. This creates a critical need to maximize the utility of limited inference-time inputs by filtering redundant observations and focusing on high-impact data. In large language models and generative artificial intelligence (AI), context engineering has emerged as a key paradigm to guide inference by selectively structuring and injecting task-relevant information. Inspired by this success, we extend context engineering to wireless systems, providing a systematic way to enhance edge AI performance without increasing model complexity. In dynamic environments, for example, beam prediction can benefit from augmenting instantaneous channel measurements with contextual cues such as user mobility trends or environment-aware propagation priors. We formally introduce wireless context engineering and propose a Wireless Context Communication Framework (WCCF) to adaptively orchestrate wireless context under inference-time constraints. This work provides researchers with a foundational perspective and practical design dimensions to manage the wireless context of wireless edge intelligence. An ISAC-enabled beam prediction case study illustrates the effectiveness of the proposed paradigm under constrained sensing budgets.
Can LLMs See Without Pixels? Benchmarking Spatial Intelligence from Textual Descriptions
Jan 07, 2026Abstract:Recent advancements in Spatial Intelligence (SI) have predominantly relied on Vision-Language Models (VLMs), yet a critical question remains: does spatial understanding originate from visual encoders or the fundamental reasoning backbone? Inspired by this question, we introduce SiT-Bench, a novel benchmark designed to evaluate the SI performance of Large Language Models (LLMs) without pixel-level input, comprises over 3,800 expert-annotated items across five primary categories and 17 subtasks, ranging from egocentric navigation and perspective transformation to fine-grained robotic manipulation. By converting single/multi-view scenes into high-fidelity, coordinate-aware textual descriptions, we challenge LLMs to perform symbolic textual reasoning rather than visual pattern matching. Evaluation results of state-of-the-art (SOTA) LLMs reveals that while models achieve proficiency in localized semantic tasks, a significant "spatial gap" remains in global consistency. Notably, we find that explicit spatial reasoning significantly boosts performance, suggesting that LLMs possess latent world-modeling potential. Our proposed dataset SiT-Bench serves as a foundational resource to foster the development of spatially-grounded LLM backbones for future VLMs and embodied agents. Our code and benchmark will be released at https://github.com/binisalegend/SiT-Bench .
EndoStreamDepth: Temporally Consistent Monocular Depth Estimation for Endoscopic Video Streams
Dec 20, 2025



Abstract:This work presents EndoStreamDepth, a monocular depth estimation framework for endoscopic video streams. It provides accurate depth maps with sharp anatomical boundaries for each frame, temporally consistent predictions across frames, and real-time throughput. Unlike prior work that uses batched inputs, EndoStreamDepth processes individual frames with a temporal module to propagate inter-frame information. The framework contains three main components: (1) a single-frame depth network with endoscopy-specific transformation to produce accurate depth maps, (2) multi-level Mamba temporal modules that leverage inter-frame information to improve accuracy and stabilize predictions, and (3) a hierarchical design with comprehensive multi-scale supervision, where complementary loss terms jointly improve local boundary sharpness and global geometric consistency. We conduct comprehensive evaluations on two publicly available colonoscopy depth estimation datasets. Compared to state-of-the-art monocular depth estimation methods, EndoStreamDepth substantially improves performance, and it produces depth maps with sharp, anatomically aligned boundaries, which are essential to support downstream tasks such as automation for robotic surgery. The code is publicly available at https://github.com/MedICL-VU/EndoStreamDepth
Agentic AI for Integrated Sensing and Communication: Analysis, Framework, and Case Study
Dec 17, 2025



Abstract:Integrated sensing and communication (ISAC) has emerged as a key development direction in the sixth-generation (6G) era, which provides essential support for the collaborative sensing and communication of future intelligent networks. However, as wireless environments become increasingly dynamic and complex, ISAC systems require more intelligent processing and more autonomous operation to maintain efficiency and adaptability. Meanwhile, agentic artificial intelligence (AI) offers a feasible solution to address these challenges by enabling continuous perception-reasoning-action loops in dynamic environments to support intelligent, autonomous, and efficient operation for ISAC systems. As such, we delve into the application value and prospects of agentic AI in ISAC systems in this work. Firstly, we provide a comprehensive review of agentic AI and ISAC systems to demonstrate their key characteristics. Secondly, we show several common optimization approaches for ISAC systems and highlight the significant advantages of generative artificial intelligence (GenAI)-based agentic AI. Thirdly, we propose a novel agentic ISAC framework and prensent a case study to verify its superiority in optimizing ISAC performance. Finally, we clarify future research directions for agentic AI-based ISAC systems.
SLMQuant:Benchmarking Small Language Model Quantization for Practical Deployment
Nov 17, 2025Abstract:Despite the growing interest in Small Language Models (SLMs) as resource-efficient alternatives to Large Language Models (LLMs), their deployment on edge devices remains challenging due to unresolved efficiency gaps in model compression. While quantization has proven effective for LLMs, its applicability to SLMs is significantly underexplored, with critical questions about differing quantization bottlenecks and efficiency profiles. This paper introduces SLMQuant, the first systematic benchmark for evaluating LLM compression techniques when applied to SLMs. Through comprehensive multi-track evaluations across diverse architectures and tasks, we analyze how state-of-the-art quantization methods perform on SLMs. Our findings reveal fundamental disparities between SLMs and LLMs in quantization sensitivity, demonstrating that direct transfer of LLM-optimized techniques leads to suboptimal results due to SLMs' unique architectural characteristics and training dynamics. We identify key factors governing effective SLM quantization and propose actionable design principles for SLM-tailored compression. SLMQuant establishes a foundational framework for advancing efficient SLM deployment on low-end devices in edge applications, and provides critical insights for deploying lightweight language models in resource-constrained scenarios.
DEMIST: \underline{DE}coupled \underline{M}ulti-stream latent d\underline{I}ffusion for Quantitative Myelin Map \underline{S}yn\underline{T}hesis
Nov 16, 2025Abstract:Quantitative magnetization transfer (qMT) imaging provides myelin-sensitive biomarkers, such as the pool size ratio (PSR), which is valuable for multiple sclerosis (MS) assessment. However, qMT requires specialized 20-30 minute scans. We propose DEMIST to synthesize PSR maps from standard T1w and FLAIR images using a 3D latent diffusion model with three complementary conditioning mechanisms. Our approach has two stages: first, we train separate autoencoders for PSR and anatomical images to learn aligned latent representations. Second, we train a conditional diffusion model in this latent space on top of a frozen diffusion foundation backbone. Conditioning is decoupled into: (i) \textbf{semantic} tokens via cross-attention, (ii) \textbf{spatial} per-scale residual hints via a 3D ControlNet branch, and (iii) \textbf{adaptive} LoRA-modulated attention. We include edge-aware loss terms to preserve lesion boundaries and alignment losses to maintain quantitative consistency, while keeping the number of trainable parameters low and retaining the inductive bias of the pretrained model. We evaluate on 163 scans from 99 subjects using 5-fold cross-validation. Our method outperforms VAE, GAN and diffusion baselines on multiple metrics, producing sharper boundaries and better quantitative agreement with ground truth. Our code is publicly available at https://github.com/MedICL-VU/MS-Synthesis-3DcLDM.
Stackelberg Game-Driven Defense for ISAC Against Channel Attacks in Low-Altitude Networks
Nov 09, 2025Abstract:The increasing saturation of terrestrial resources has driven economic activities into low-altitude airspace. These activities, such as air taxis, rely on low-altitude wireless networks, and one key enabling technology is integrated sensing and communication (ISAC). However, in low-altitude airspace, ISAC is vulnerable to channel-access attacks, thereby degrading performance and threatening safety. To address this, we propose a defense framework based on a Stackelberg game. Specifically, we first model the system under attack, deriving metrics for the communication and the sensing to quantify performance. Then, we formulate the interaction as a three-player game where a malicious attacker acts as the leader, while the legitimate drone and ground base station act as followers. Using a backward induction algorithm, we obtain the Stackelberg equilibrium, allowing the defenders to dynamically adjust their strategies to mitigate the attack. Simulation results verify that the proposed algorithm converges to a stable solution and outperforms existing baselines, ensuring reliable ISAC performance for critical low-altitude applications.
Low-Altitude UAV-Carried Movable Antenna for Joint Wireless Power Transfer and Covert Communications
Oct 30, 2025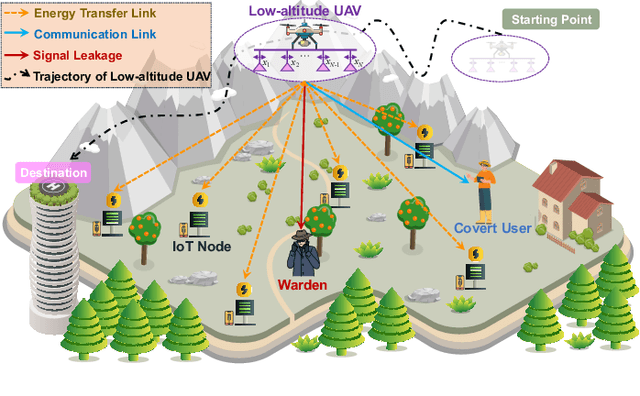
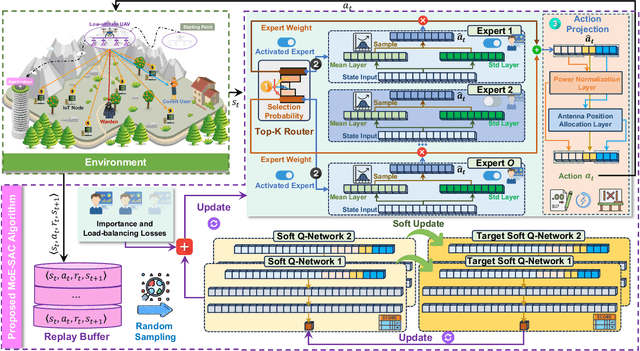
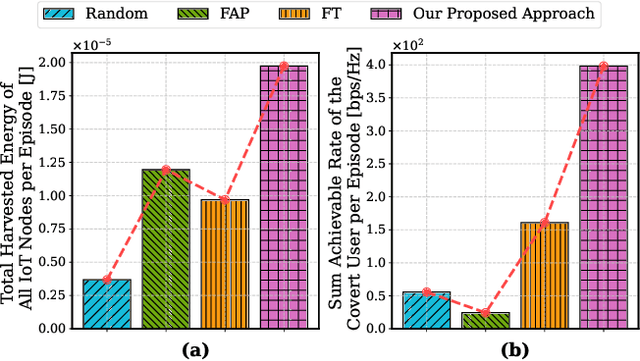
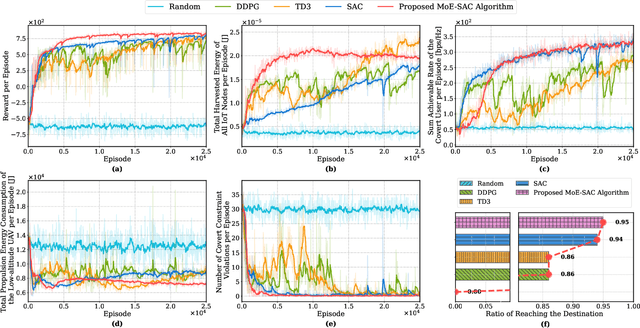
Abstract:The proliferation of Internet of Things (IoT) networks has created an urgent need for sustainable energy solutions, particularly for the battery-constrained spatially distributed IoT nodes. While low-altitude uncrewed aerial vehicles (UAVs) employed with wireless power transfer (WPT) capabilities offer a promising solution, the line-of-sight channels that facilitate efficient energy delivery also expose sensitive operational data to adversaries. This paper proposes a novel low-altitude UAV-carried movable antenna-enhanced transmission system joint WPT and covert communications, which simultaneously performs energy supplements to IoT nodes and establishes transmission links with a covert user by leveraging wireless energy signals as a natural cover. Then, we formulate a multi-objective optimization problem that jointly maximizes the total harvested energy of IoT nodes and sum achievable rate of the covert user, while minimizing the propulsion energy consumption of the low-altitude UAV. To address the non-convex and temporally coupled optimization problem, we propose a mixture-of-experts-augmented soft actor-critic (MoE-SAC) algorithm that employs a sparse Top-K gated mixture-of-shallow-experts architecture to represent multimodal policy distributions arising from the conflicting optimization objectives. We also incorporate an action projection module that explicitly enforces per-time-slot power budget constraints and antenna position constraints. Simulation results demonstrate that the proposed approach significantly outperforms some baseline approaches and other state-of-the-art deep reinforcement learning algorithms.
Cost Minimization for Space-Air-Ground Integrated Multi-Access Edge Computing Systems
Oct 24, 2025Abstract:Space-air-ground integrated multi-access edge computing (SAGIN-MEC) provides a promising solution for the rapidly developing low-altitude economy (LAE) to deliver flexible and wide-area computing services. However, fully realizing the potential of SAGIN-MEC in the LAE presents significant challenges, including coordinating decisions across heterogeneous nodes with different roles, modeling complex factors such as mobility and network variability, and handling real-time decision-making under partially observable environment with hybrid variables. To address these challenges, we first present a hierarchical SAGIN-MEC architecture that enables the coordination between user devices (UDs), uncrewed aerial vehicles (UAVs), and satellites. Then, we formulate a UD cost minimization optimization problem (UCMOP) to minimize the UD cost by jointly optimizing the task offloading ratio, UAV trajectory planning, computing resource allocation, and UD association. We show that the UCMOP is an NP-hard problem. To overcome this challenge, we propose a multi-agent deep deterministic policy gradient (MADDPG)-convex optimization and coalitional game (MADDPG-COCG) algorithm. Specifically, we employ the MADDPG algorithm to optimize the continuous temporal decisions for heterogeneous nodes in the partially observable SAGIN-MEC system. Moreover, we propose a convex optimization and coalitional game (COCG) method to enhance the conventional MADDPG by deterministically handling the hybrid and varying-dimensional decisions. Simulation results demonstrate that the proposed MADDPG-COCG algorithm significantly enhances the user-centric performances in terms of the aggregated UD cost, task completion delay, and UD energy consumption, with a slight increase in UAV energy consumption, compared to the benchmark algorithms. Moreover, the MADDPG-COCG algorithm shows superior convergence stability and scalability.
Joint AoI and Handover Optimization in Space-Air-Ground Integrated Network
Sep 16, 2025Abstract:Despite the widespread deployment of terrestrial networks, providing reliable communication services to remote areas and maintaining connectivity during emergencies remains challenging. Low Earth orbit (LEO) satellite constellations offer promising solutions with their global coverage capabilities and reduced latency, yet struggle with intermittent coverage and limited communication windows due to orbital dynamics. This paper introduces an age of information (AoI)-aware space-air-ground integrated network (SAGIN) architecture that leverages a high-altitude platform (HAP) as intelligent relay between the LEO satellites and ground terminals. Our three-layer design employs hybrid free-space optical (FSO) links for high-capacity satellite-to-HAP communication and reliable radio frequency (RF) links for HAP-to-ground transmission, and thus addressing the temporal discontinuity in LEO satellite coverage while serving diverse user priorities. Specifically, we formulate a joint optimization problem to simultaneously minimize the AoI and satellite handover frequency through optimal transmit power distribution and satellite selection decisions. This highly dynamic, non-convex problem with time-coupled constraints presents significant computational challenges for traditional approaches. To address these difficulties, we propose a novel diffusion model (DM)-enhanced dueling double deep Q-network with action decomposition and state transformer encoder (DD3QN-AS) algorithm that incorporates transformer-based temporal feature extraction and employs a DM-based latent prompt generative module to refine state-action representations through conditional denoising. Simulation results highlight the superior performance of the proposed approach compared with policy-based methods and some other deep reinforcement learning (DRL) benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge