Michael M. Hoffman
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022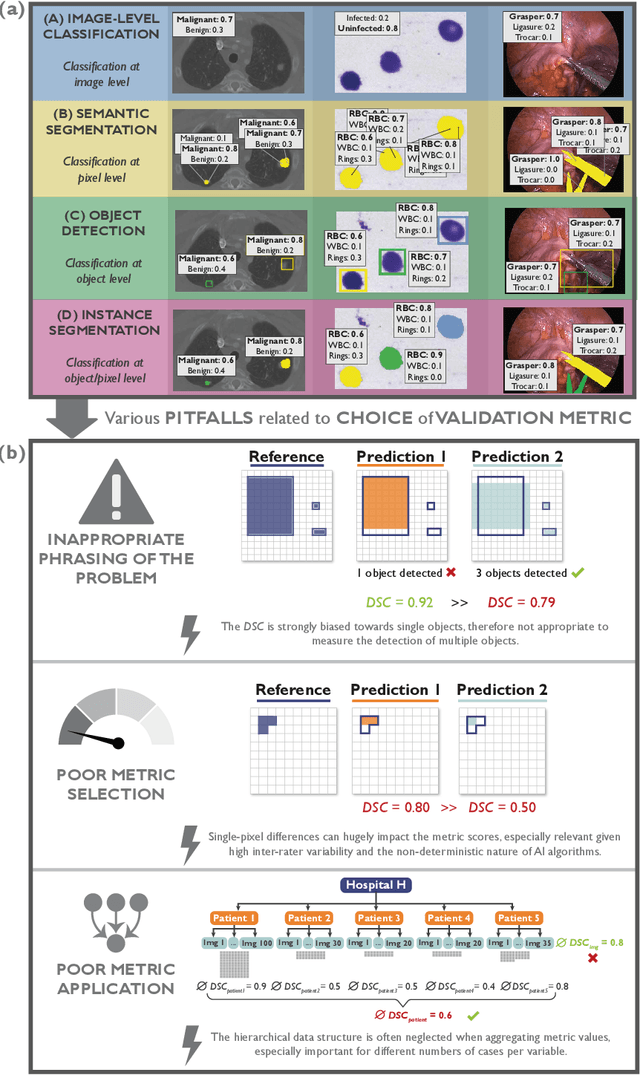
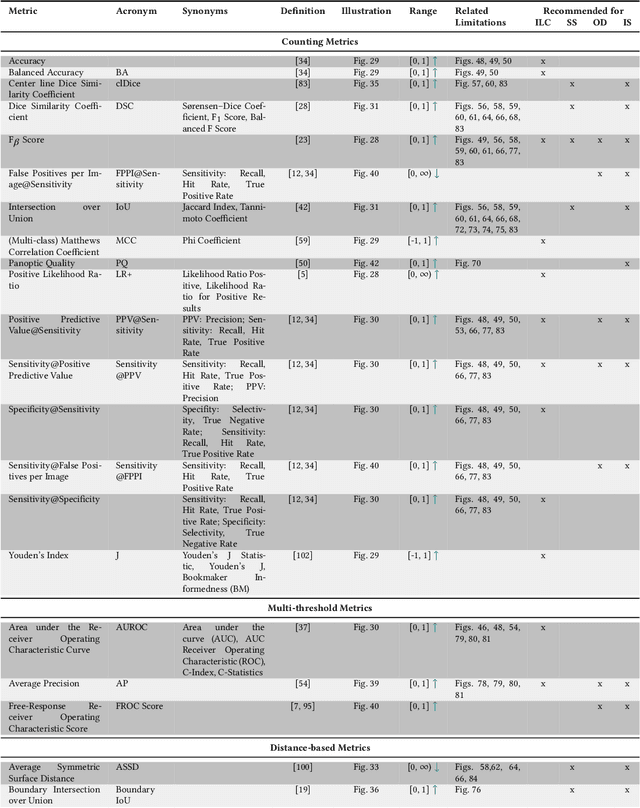
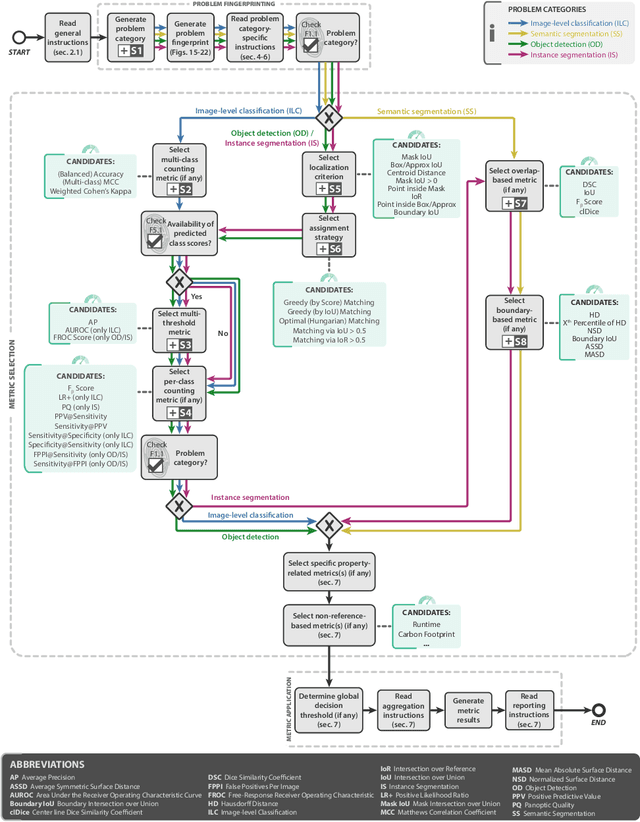
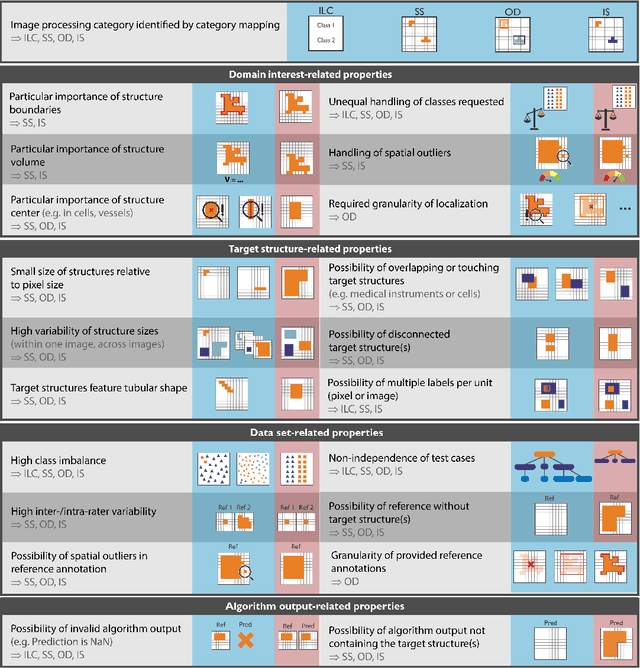
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
The MCC-F1 curve: a performance evaluation technique for binary classification
Jun 17, 2020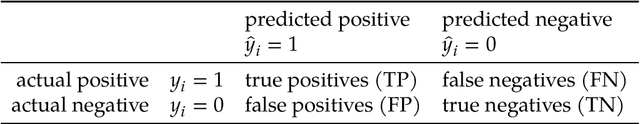
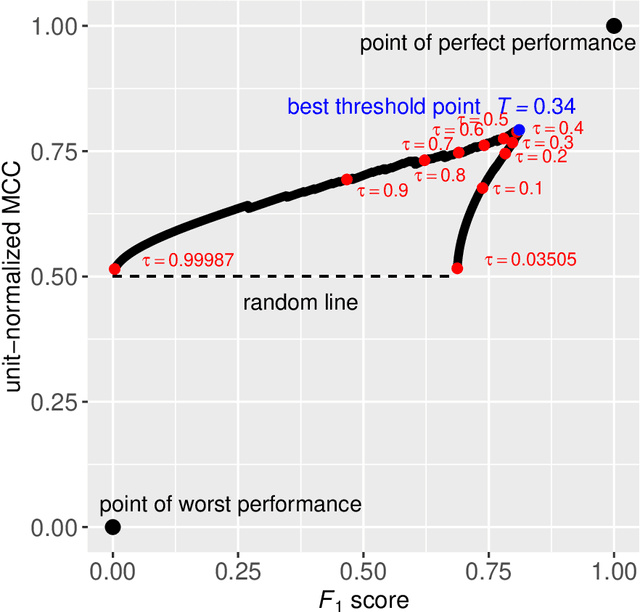
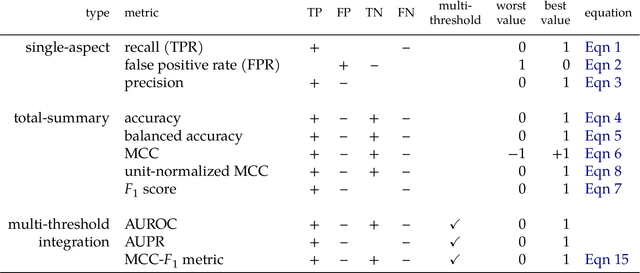
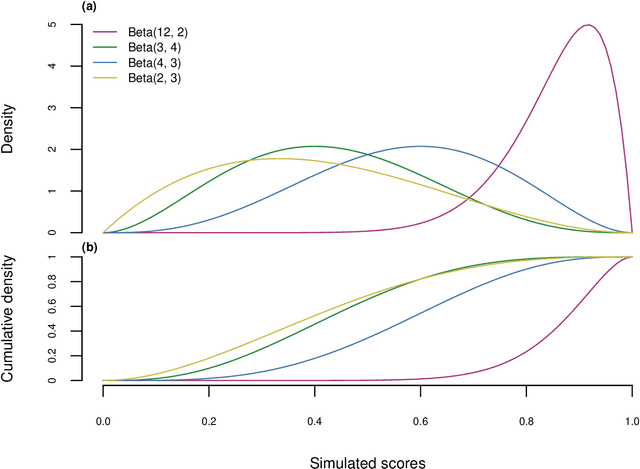
Abstract:Many fields use the ROC curve and the PR curve as standard evaluations of binary classification methods. Analysis of ROC and PR, however, often gives misleading and inflated performance evaluations, especially with an imbalanced ground truth. Here, we demonstrate the problems with ROC and PR analysis through simulations, and propose the MCC-F1 curve to address these drawbacks. The MCC-F1 curve combines two informative single-threshold metrics, MCC and the F1 score. The MCC-F1 curve more clearly differentiates good and bad classifiers, even with imbalanced ground truths. We also introduce the MCC-F1 metric, which provides a single value that integrates many aspects of classifier performance across the whole range of classification thresholds. Finally, we provide an R package that plots MCC-F1 curves and calculates related metrics.
Machine Learning for Integrating Data in Biology and Medicine: Principles, Practice, and Opportunities
Oct 10, 2018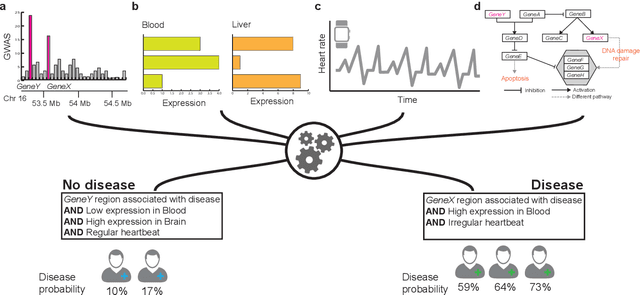
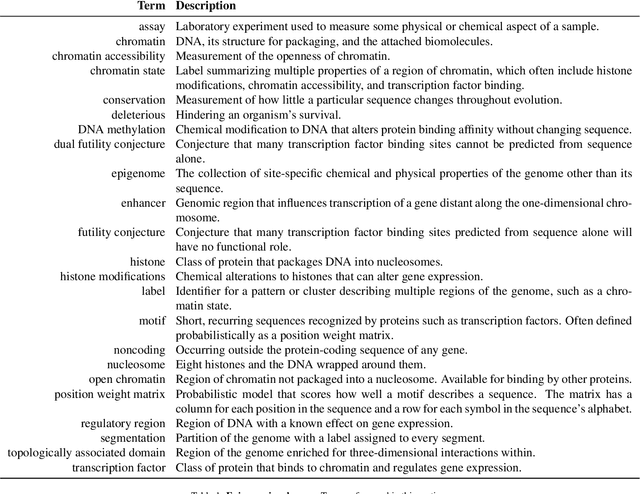
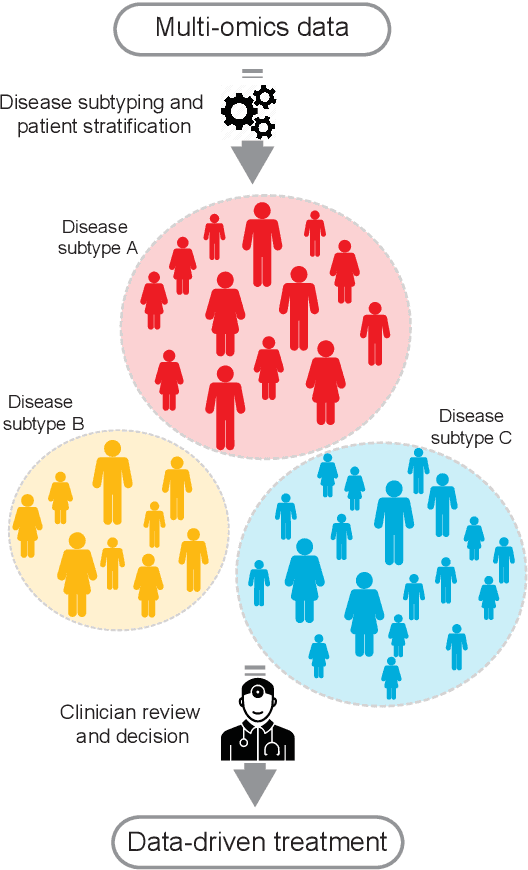
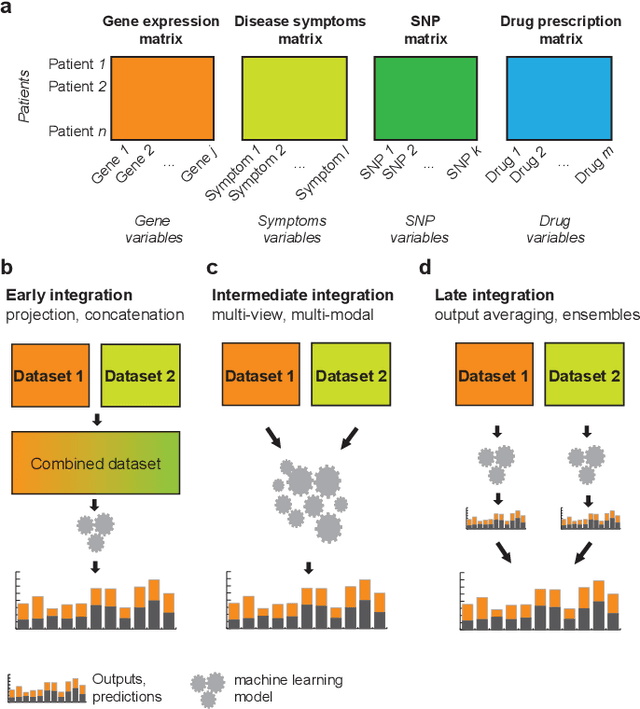
Abstract:New technologies have enabled the investigation of biology and human health at an unprecedented scale and in multiple dimensions. These dimensions include a myriad of properties describing genome, epigenome, transcriptome, microbiome, phenotype, and lifestyle. No single data type, however, can capture the complexity of all the factors relevant to understanding a phenomenon such as a disease. Integrative methods that combine data from multiple technologies have thus emerged as critical statistical and computational approaches. The key challenge in developing such approaches is the identification of effective models to provide a comprehensive and relevant systems view. An ideal method can answer a biological or medical question, identifying important features and predicting outcomes, by harnessing heterogeneous data across several dimensions of biological variation. In this Review, we describe the principles of data integration and discuss current methods and available implementations. We provide examples of successful data integration in biology and medicine. Finally, we discuss current challenges in biomedical integrative methods and our perspective on the future development of the field.
Statistical Inference, Learning and Models in Big Data
Jan 28, 2016
Abstract:The need for new methods to deal with big data is a common theme in most scientific fields, although its definition tends to vary with the context. Statistical ideas are an essential part of this, and as a partial response, a thematic program on statistical inference, learning, and models in big data was held in 2015 in Canada, under the general direction of the Canadian Statistical Sciences Institute, with major funding from, and most activities located at, the Fields Institute for Research in Mathematical Sciences. This paper gives an overview of the topics covered, describing challenges and strategies that seem common to many different areas of application, and including some examples of applications to make these challenges and strategies more concrete.
* Thematic Program on Statistical Inference, Learning, and Models for Big Data, Fields Institute; 23 pages, 2 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge