Bernhard Kainz
Biomedical Image Analysis Group, Department of Computing, Imperial College London
ShapePuri: Shape Guided and Appearance Generalized Adversarial Purification
Feb 05, 2026Abstract:Deep neural networks demonstrate impressive performance in visual recognition, but they remain vulnerable to adversarial attacks that is imperceptible to the human. Although existing defense strategies such as adversarial training and purification have achieved progress, diffusion-based purification often involves high computational costs and information loss. To address these challenges, we introduce Shape Guided Purification (ShapePuri), a novel defense framework enhances robustness by aligning model representations with stable structural invariants. ShapePuri integrates two components: a Shape Encoding Module (SEM) that provides dense geometric guidance through Signed Distance Functions (SDF), and a Global Appearance Debiasing (GAD) module that mitigates appearance bias via stochastic transformations. In our experiments, ShapePuri achieves $84.06\%$ clean accuracy and $81.64\%$ robust accuracy under the AutoAttack protocol, representing the first defense framework to surpass the $80\%$ threshold on this benchmark. Our approach provides a scalable and efficient adversarial defense that preserves prediction stability during inference without requiring auxiliary modules or additional computational cost.
Measuring and Aligning Abstraction in Vision-Language Models with Medical Taxonomies
Jan 21, 2026Abstract:Vision-Language Models show strong zero-shot performance for chest X-ray classification, but standard flat metrics fail to distinguish between clinically minor and severe errors. This work investigates how to quantify and mitigate abstraction errors by leveraging medical taxonomies. We benchmark several state-of-the-art VLMs using hierarchical metrics and introduce Catastrophic Abstraction Errors to capture cross-branch mistakes. Our results reveal substantial misalignment of VLMs with clinical taxonomies despite high flat performance. To address this, we propose risk-constrained thresholding and taxonomy-aware fine-tuning with radial embeddings, which reduce severe abstraction errors to below 2 per cent while maintaining competitive performance. These findings highlight the importance of hierarchical evaluation and representation-level alignment for safer and more clinically meaningful deployment of VLMs.
LCMem: A Universal Model for Robust Image Memorization Detection
Dec 16, 2025Abstract:Recent advances in generative image modeling have achieved visual realism sufficient to deceive human experts, yet their potential for privacy preserving data sharing remains insufficiently understood. A central obstacle is the absence of reliable memorization detection mechanisms, limited quantitative evaluation, and poor generalization of existing privacy auditing methods across domains. To address this, we propose to view memorization detection as a unified problem at the intersection of re-identification and copy detection, whose complementary goals cover both identity consistency and augmentation-robust duplication, and introduce Latent Contrastive Memorization Network (LCMem), a cross-domain model evaluated jointly on both tasks. LCMem achieves this through a two-stage training strategy that first learns identity consistency before incorporating augmentation-robust copy detection. Across six benchmark datasets, LCMem achieves improvements of up to 16 percentage points on re-identification and 30 percentage points on copy detection, enabling substantially more reliable memorization detection at scale. Our results show that existing privacy filters provide limited performance and robustness, highlighting the need for stronger protection mechanisms. We show that LCMem sets a new standard for cross-domain privacy auditing, offering reliable and scalable memorization detection. Code and model is publicly available at https://github.com/MischaD/LCMem.
STARCaster: Spatio-Temporal AutoRegressive Video Diffusion for Identity- and View-Aware Talking Portraits
Dec 15, 2025



Abstract:This paper presents STARCaster, an identity-aware spatio-temporal video diffusion model that addresses both speech-driven portrait animation and free-viewpoint talking portrait synthesis, given an identity embedding or reference image, within a unified framework. Existing 2D speech-to-video diffusion models depend heavily on reference guidance, leading to limited motion diversity. At the same time, 3D-aware animation typically relies on inversion through pre-trained tri-plane generators, which often leads to imperfect reconstructions and identity drift. We rethink reference- and geometry-based paradigms in two ways. First, we deviate from strict reference conditioning at pre-training by introducing softer identity constraints. Second, we address 3D awareness implicitly within the 2D video domain by leveraging the inherent multi-view nature of video data. STARCaster adopts a compositional approach progressing from ID-aware motion modeling, to audio-visual synchronization via lip reading-based supervision, and finally to novel view animation through temporal-to-spatial adaptation. To overcome the scarcity of 4D audio-visual data, we propose a decoupled learning approach in which view consistency and temporal coherence are trained independently. A self-forcing training scheme enables the model to learn from longer temporal contexts than those generated at inference, mitigating the overly static animations common in existing autoregressive approaches. Comprehensive evaluations demonstrate that STARCaster generalizes effectively across tasks and identities, consistently surpassing prior approaches in different benchmarks.
InfoMotion: A Graph-Based Approach to Video Dataset Distillation for Echocardiography
Dec 13, 2025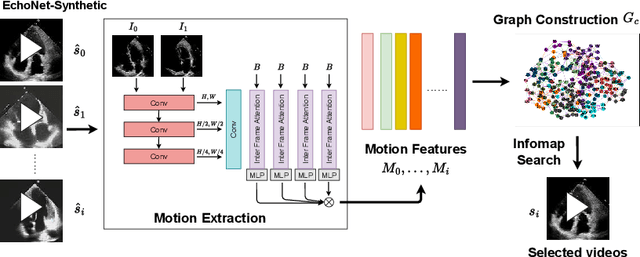
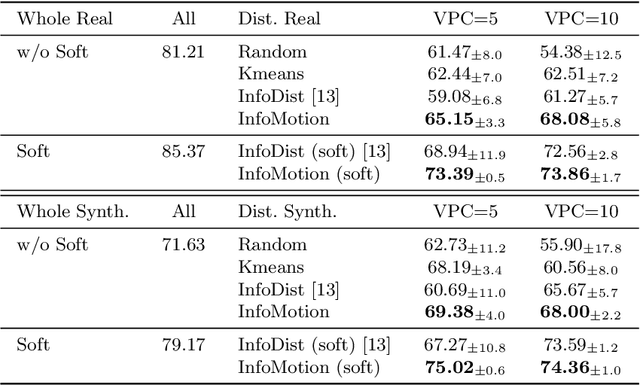
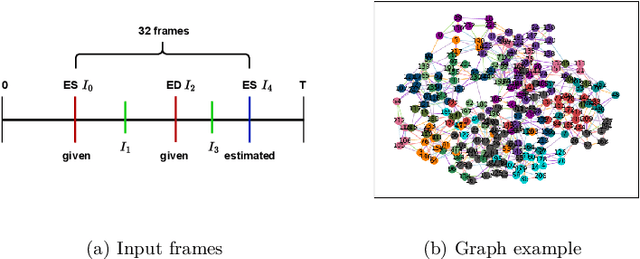
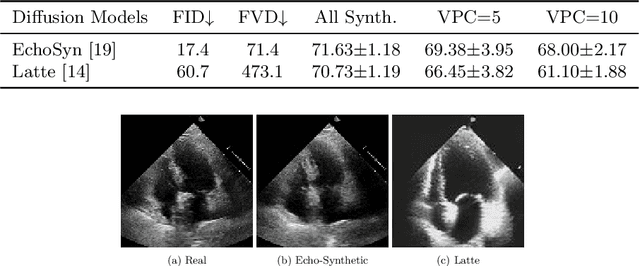
Abstract:Echocardiography plays a critical role in the diagnosis and monitoring of cardiovascular diseases as a non-invasive real-time assessment of cardiac structure and function. However, the growing scale of echocardiographic video data presents significant challenges in terms of storage, computation, and model training efficiency. Dataset distillation offers a promising solution by synthesizing a compact, informative subset of data that retains the key clinical features of the original dataset. In this work, we propose a novel approach for distilling a compact synthetic echocardiographic video dataset. Our method leverages motion feature extraction to capture temporal dynamics, followed by class-wise graph construction and representative sample selection using the Infomap algorithm. This enables us to select a diverse and informative subset of synthetic videos that preserves the essential characteristics of the original dataset. We evaluate our approach on the EchoNet-Dynamic datasets and achieve a test accuracy of \(69.38\%\) using only \(25\) synthetic videos. These results demonstrate the effectiveness and scalability of our method for medical video dataset distillation.
Label-free Motion-Conditioned Diffusion Model for Cardiac Ultrasound Synthesis
Dec 10, 2025Abstract:Ultrasound echocardiography is essential for the non-invasive, real-time assessment of cardiac function, but the scarcity of labelled data, driven by privacy restrictions and the complexity of expert annotation, remains a major obstacle for deep learning methods. We propose the Motion Conditioned Diffusion Model (MCDM), a label-free latent diffusion framework that synthesises realistic echocardiography videos conditioned on self-supervised motion features. To extract these features, we design the Motion and Appearance Feature Extractor (MAFE), which disentangles motion and appearance representations from videos. Feature learning is further enhanced by two auxiliary objectives: a re-identification loss guided by pseudo appearance features and an optical flow loss guided by pseudo flow fields. Evaluated on the EchoNet-Dynamic dataset, MCDM achieves competitive video generation performance, producing temporally coherent and clinically realistic sequences without reliance on manual labels. These results demonstrate the potential of self-supervised conditioning for scalable echocardiography synthesis. Our code is available at https://github.com/ZheLi2020/LabelfreeMCDM.
Better Tokens for Better 3D: Advancing Vision-Language Modeling in 3D Medical Imaging
Oct 23, 2025Abstract:Recent progress in vision-language modeling for 3D medical imaging has been fueled by large-scale computed tomography (CT) corpora with paired free-text reports, stronger architectures, and powerful pretrained models. This has enabled applications such as automated report generation and text-conditioned 3D image synthesis. Yet, current approaches struggle with high-resolution, long-sequence volumes: contrastive pretraining often yields vision encoders that are misaligned with clinical language, and slice-wise tokenization blurs fine anatomy, reducing diagnostic performance on downstream tasks. We introduce BTB3D (Better Tokens for Better 3D), a causal convolutional encoder-decoder that unifies 2D and 3D training and inference while producing compact, frequency-aware volumetric tokens. A three-stage training curriculum enables (i) local reconstruction, (ii) overlapping-window tiling, and (iii) long-context decoder refinement, during which the model learns from short slice excerpts yet generalizes to scans exceeding 300 slices without additional memory overhead. BTB3D sets a new state-of-the-art on two key tasks: it improves BLEU scores and increases clinical F1 by 40% over CT2Rep, CT-CHAT, and Merlin for report generation; and it reduces FID by 75% and halves FVD compared to GenerateCT and MedSyn for text-to-CT synthesis, producing anatomically consistent 512*512*241 volumes. These results confirm that precise three-dimensional tokenization, rather than larger language backbones alone, is essential for scalable vision-language modeling in 3D medical imaging. The codebase is available at: https://github.com/ibrahimethemhamamci/BTB3D
Graph Conditioned Diffusion for Controllable Histopathology Image Generation
Oct 08, 2025Abstract:Recent advances in Diffusion Probabilistic Models (DPMs) have set new standards in high-quality image synthesis. Yet, controlled generation remains challenging, particularly in sensitive areas such as medical imaging. Medical images feature inherent structure such as consistent spatial arrangement, shape or texture, all of which are critical for diagnosis. However, existing DPMs operate in noisy latent spaces that lack semantic structure and strong priors, making it difficult to ensure meaningful control over generated content. To address this, we propose graph-based object-level representations for Graph-Conditioned-Diffusion. Our approach generates graph nodes corresponding to each major structure in the image, encapsulating their individual features and relationships. These graph representations are processed by a transformer module and integrated into a diffusion model via the text-conditioning mechanism, enabling fine-grained control over generation. We evaluate this approach using a real-world histopathology use case, demonstrating that our generated data can reliably substitute for annotated patient data in downstream segmentation tasks. The code is available here.
Ontology-Based Concept Distillation for Radiology Report Retrieval and Labeling
Aug 27, 2025Abstract:Retrieval-augmented learning based on radiology reports has emerged as a promising direction to improve performance on long-tail medical imaging tasks, such as rare disease detection in chest X-rays. Most existing methods rely on comparing high-dimensional text embeddings from models like CLIP or CXR-BERT, which are often difficult to interpret, computationally expensive, and not well-aligned with the structured nature of medical knowledge. We propose a novel, ontology-driven alternative for comparing radiology report texts based on clinically grounded concepts from the Unified Medical Language System (UMLS). Our method extracts standardised medical entities from free-text reports using an enhanced pipeline built on RadGraph-XL and SapBERT. These entities are linked to UMLS concepts (CUIs), enabling a transparent, interpretable set-based representation of each report. We then define a task-adaptive similarity measure based on a modified and weighted version of the Tversky Index that accounts for synonymy, negation, and hierarchical relationships between medical entities. This allows efficient and semantically meaningful similarity comparisons between reports. We demonstrate that our approach outperforms state-of-the-art embedding-based retrieval methods in a radiograph classification task on MIMIC-CXR, particularly in long-tail settings. Additionally, we use our pipeline to generate ontology-backed disease labels for MIMIC-CXR, offering a valuable new resource for downstream learning tasks. Our work provides more explainable, reliable, and task-specific retrieval strategies in clinical AI systems, especially when interpretability and domain knowledge integration are essential. Our code is available at https://github.com/Felix-012/ontology-concept-distillation
Diffusing the Blind Spot: Uterine MRI Synthesis with Diffusion Models
Aug 11, 2025


Abstract:Despite significant progress in generative modelling, existing diffusion models often struggle to produce anatomically precise female pelvic images, limiting their application in gynaecological imaging, where data scarcity and patient privacy concerns are critical. To overcome these barriers, we introduce a novel diffusion-based framework for uterine MRI synthesis, integrating both unconditional and conditioned Denoising Diffusion Probabilistic Models (DDPMs) and Latent Diffusion Models (LDMs) in 2D and 3D. Our approach generates anatomically coherent, high fidelity synthetic images that closely mimic real scans and provide valuable resources for training robust diagnostic models. We evaluate generative quality using advanced perceptual and distributional metrics, benchmarking against standard reconstruction methods, and demonstrate substantial gains in diagnostic accuracy on a key classification task. A blinded expert evaluation further validates the clinical realism of our synthetic images. We release our models with privacy safeguards and a comprehensive synthetic uterine MRI dataset to support reproducible research and advance equitable AI in gynaecology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge