A. Emre Kavur
Medical Imaging AI Competitions Lack Fairness
Dec 19, 2025



Abstract:Benchmarking competitions are central to the development of artificial intelligence (AI) in medical imaging, defining performance standards and shaping methodological progress. However, it remains unclear whether these benchmarks provide data that are sufficiently representative, accessible, and reusable to support clinically meaningful AI. In this work, we assess fairness along two complementary dimensions: (1) whether challenge datasets are representative of real-world clinical diversity, and (2) whether they are accessible and legally reusable in line with the FAIR principles. To address this question, we conducted a large-scale systematic study of 241 biomedical image analysis challenges comprising 458 tasks across 19 imaging modalities. Our findings show substantial biases in dataset composition, including geographic location, modality-, and problem type-related biases, indicating that current benchmarks do not adequately reflect real-world clinical diversity. Despite their widespread influence, challenge datasets were frequently constrained by restrictive or ambiguous access conditions, inconsistent or non-compliant licensing practices, and incomplete documentation, limiting reproducibility and long-term reuse. Together, these shortcomings expose foundational fairness limitations in our benchmarking ecosystem and highlight a disconnect between leaderboard success and clinical relevance.
Bridging vision language model (VLM) evaluation gaps with a framework for scalable and cost-effective benchmark generation
Feb 21, 2025



Abstract:Reliable evaluation of AI models is critical for scientific progress and practical application. While existing VLM benchmarks provide general insights into model capabilities, their heterogeneous designs and limited focus on a few imaging domains pose significant challenges for both cross-domain performance comparison and targeted domain-specific evaluation. To address this, we propose three key contributions: (1) a framework for the resource-efficient creation of domain-specific VLM benchmarks enabled by task augmentation for creating multiple diverse tasks from a single existing task, (2) the release of new VLM benchmarks for seven domains, created according to the same homogeneous protocol and including 162,946 thoroughly human-validated answers, and (3) an extensive benchmarking of 22 state-of-the-art VLMs on a total of 37,171 tasks, revealing performance variances across domains and tasks, thereby supporting the need for tailored VLM benchmarks. Adoption of our methodology will pave the way for the resource-efficient domain-specific selection of models and guide future research efforts toward addressing core open questions.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Labeling instructions matter in biomedical image analysis
Jul 20, 2022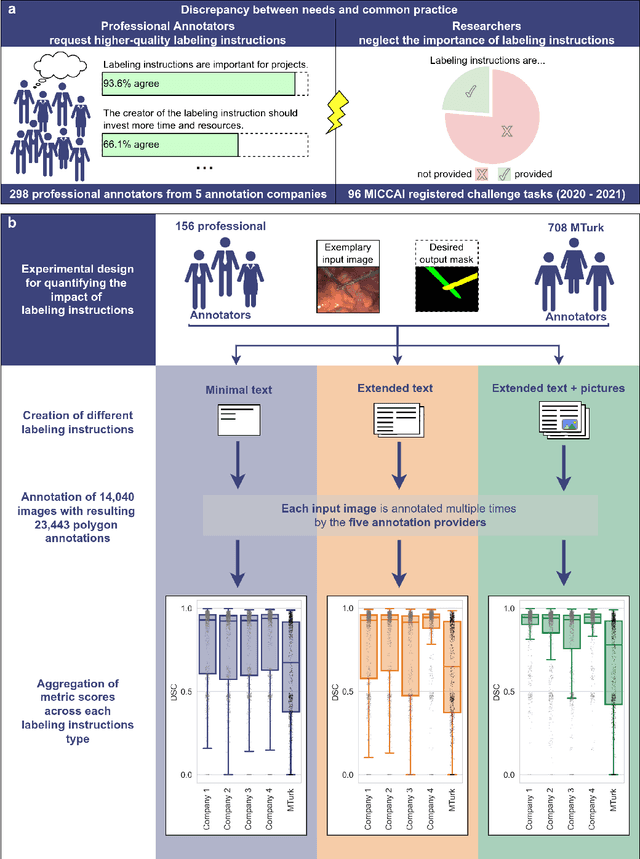
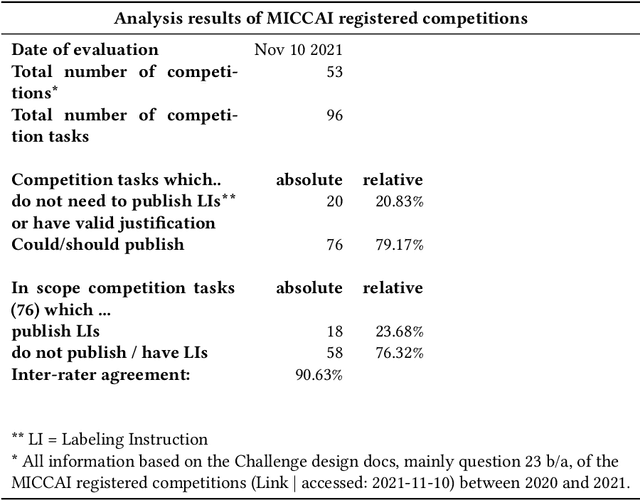
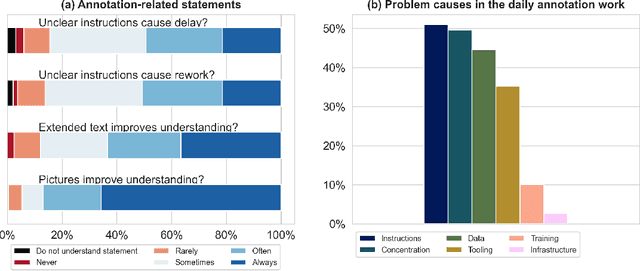
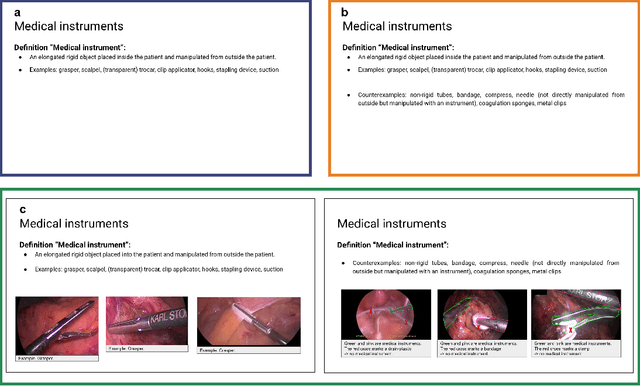
Abstract:Biomedical image analysis algorithm validation depends on high-quality annotation of reference datasets, for which labeling instructions are key. Despite their importance, their optimization remains largely unexplored. Here, we present the first systematic study of labeling instructions and their impact on annotation quality in the field. Through comprehensive examination of professional practice and international competitions registered at the MICCAI Society, we uncovered a discrepancy between annotators' needs for labeling instructions and their current quality and availability. Based on an analysis of 14,040 images annotated by 156 annotators from four professional companies and 708 Amazon Mechanical Turk (MTurk) crowdworkers using instructions with different information density levels, we further found that including exemplary images significantly boosts annotation performance compared to text-only descriptions, while solely extending text descriptions does not. Finally, professional annotators constantly outperform MTurk crowdworkers. Our study raises awareness for the need of quality standards in biomedical image analysis labeling instructions.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022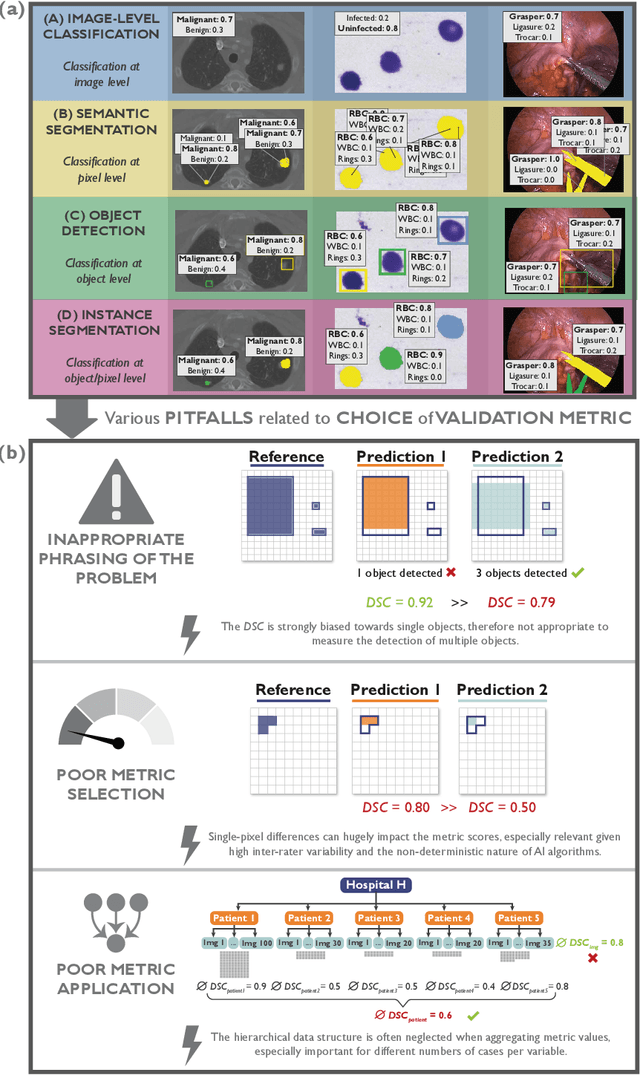
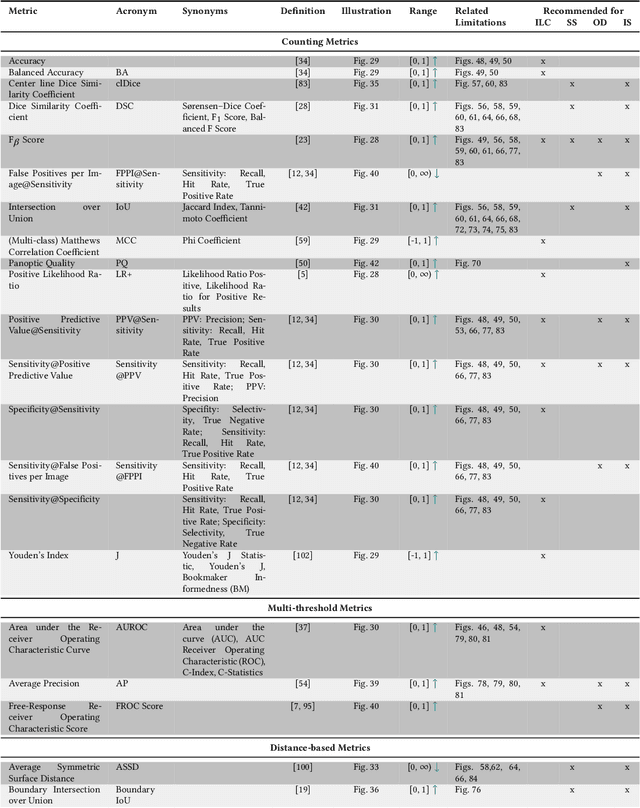
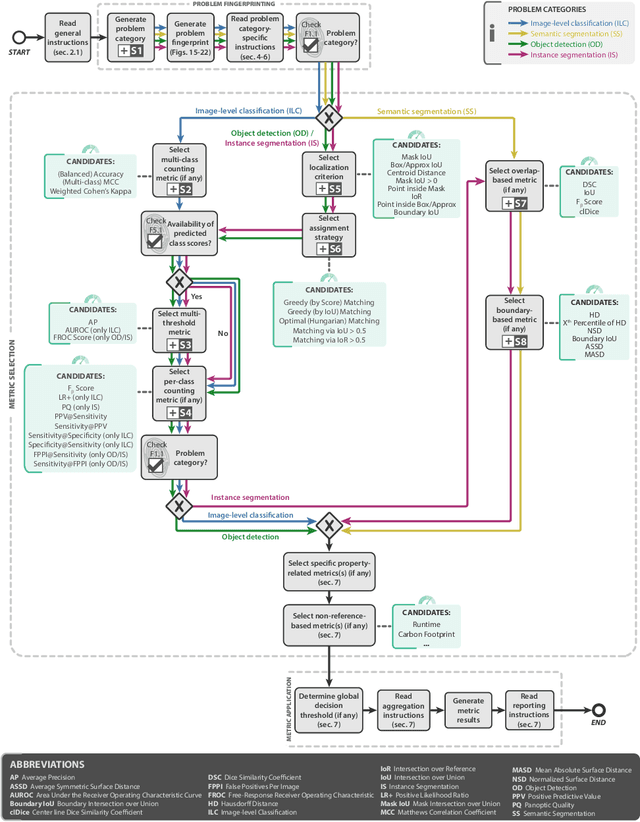
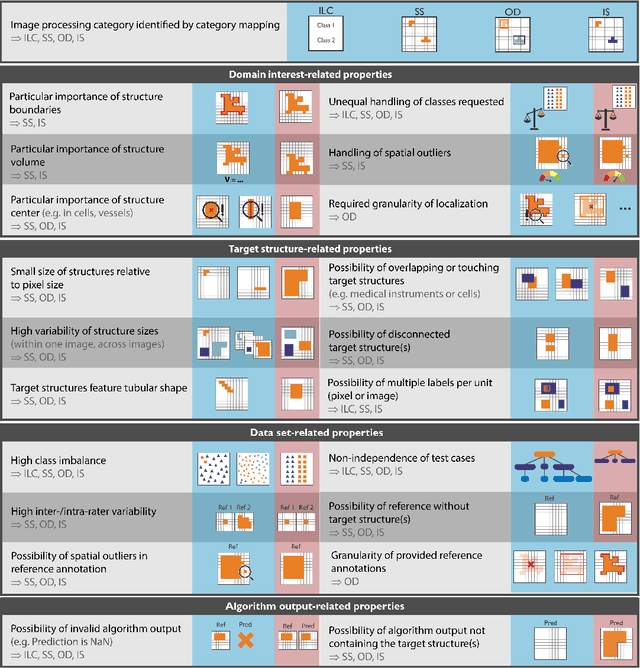
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
Cross-Domain Segmentation with Adversarial Loss and Covariate Shift for Biomedical Imaging
Jun 08, 2020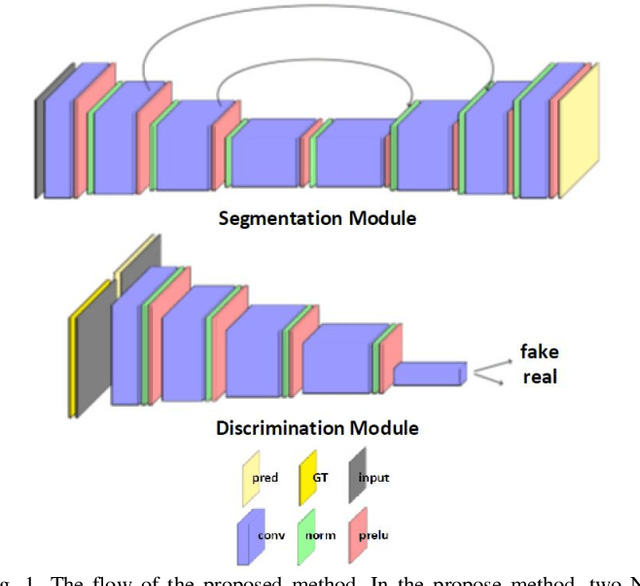
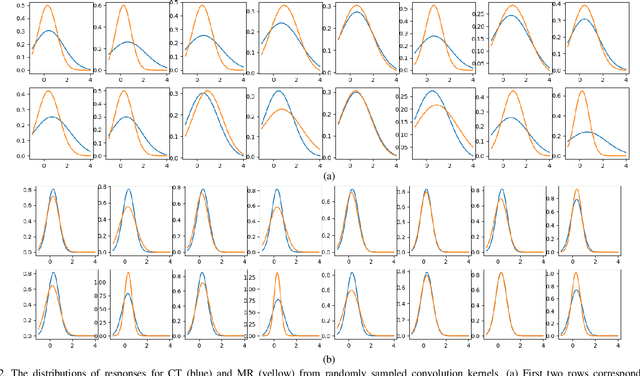
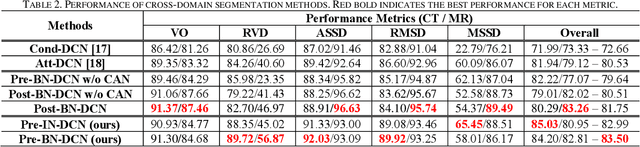
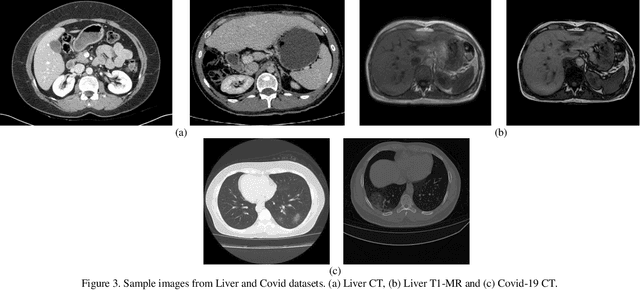
Abstract:Despite the widespread use of deep learning methods for semantic segmentation of images that are acquired from a single source, clinicians often use multi-domain data for a detailed analysis. For instance, CT and MRI have advantages over each other in terms of imaging quality, artifacts, and output characteristics that lead to differential diagnosis. The capacity of current segmentation techniques is only allow to work for an individual domain due to their differences. However, the models that are capable of working on all modalities are essentially needed for a complete solution. Furthermore, robustness is drastically affected by the number of samples in the training step, especially for deep learning models. Hence, there is a necessity that all available data regardless of data domain should be used for reliable methods. For this purpose, this manuscript aims to implement a novel model that can learn robust representations from cross-domain data by encapsulating distinct and shared patterns from different modalities. Precisely, covariate shift property is retained with structural modification and adversarial loss where sparse and rich representations are obtained. Hence, a single parameter set is used to perform cross-domain segmentation task. The superiority of the proposed method is that no information related to modalities are provided in either training or inference phase. The tests on CT and MRI liver data acquired in routine clinical workflows show that the proposed model outperforms all other baseline with a large margin. Experiments are also conducted on Covid-19 dataset that it consists of CT data where significant intra-class visual differences are observed. Similarly, the proposed method achieves the best performance.
CHAOS Challenge -- Combined (CT-MR) Healthy Abdominal Organ Segmentation
Jan 17, 2020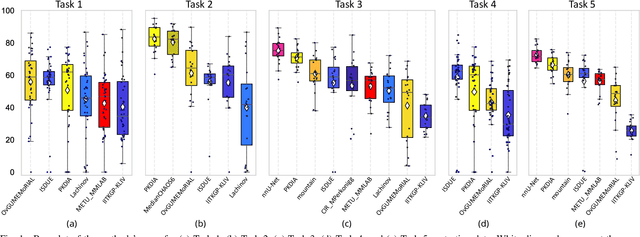
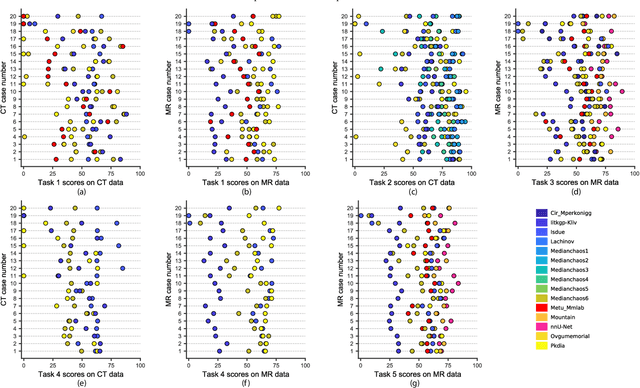
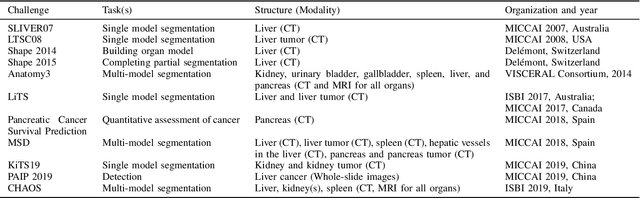
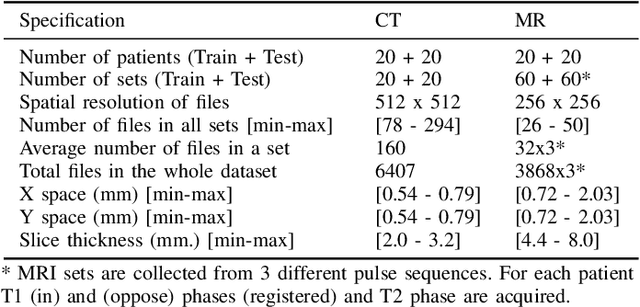
Abstract:Segmentation of abdominal organs has been a comprehensive, yet unresolved, research field for many years. In the last decade, intensive developments in deep learning (DL) have introduced new state-of-the-art segmentation systems. Despite outperforming the overall accuracy of existing systems, the effects of DL model properties and parameters on the performance is hard to interpret. This makes comparative analysis a necessary tool to achieve explainable studies and systems. Moreover, the performance of DL for emerging learning approaches such as cross-modality and multi-modal tasks have been rarely discussed. In order to expand the knowledge in these topics, CHAOS -- Combined (CT-MR) Healthy Abdominal Organ Segmentation challenge has been organized in the IEEE International Symposium on Biomedical Imaging (ISBI), 2019, in Venice, Italy. Despite a large number of the previous abdomen related challenges, the majority of which are focused on tumor/lesion detection and/or classification with a single modality, CHAOS provides both abdominal CT and MR data from healthy subjects. Five different and complementary tasks have been designed to analyze the capabilities of the current approaches from multiple perspectives. The results are investigated thoroughly, compared with manual annotations and interactive methods. The outcomes are reported in detail to reflect the latest advancements in the field. CHAOS challenge and data will be available online to provide a continuous benchmark resource for segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge