Sebastien Ourselin
King's College London
From Pre- to Intra-operative MRI: Predicting Brain Shift in Temporal Lobe Resection for Epilepsy Surgery
Feb 03, 2026Abstract:Introduction: In neurosurgery, image-guided Neurosurgery Systems (IGNS) highly rely on preoperative brain magnetic resonance images (MRI) to assist surgeons in locating surgical targets and determining surgical paths. However, brain shift invalidates the preoperative MRI after dural opening. Updated intraoperative brain MRI with brain shift compensation is crucial for enhancing the precision of neuronavigation systems and ensuring the optimal outcome of surgical interventions. Methodology: We propose NeuralShift, a U-Net-based model that predicts brain shift entirely from pre-operative MRI for patients undergoing temporal lobe resection. We evaluated our results using Target Registration Errors (TREs) computed on anatomical landmarks located on the resection side and along the midline, and DICE scores comparing predicted intraoperative masks with masks derived from intraoperative MRI. Results: Our experimental results show that our model can predict the global deformation of the brain (DICE of 0.97) with accurate local displacements (achieve landmark TRE as low as 1.12 mm), compensating for large brain shifts during temporal lobe removal neurosurgery. Conclusion: Our proposed model is capable of predicting the global deformation of the brain during temporal lobe resection using only preoperative images, providing potential opportunities to the surgical team to increase safety and efficiency of neurosurgery and better outcomes to patients. Our contributions will be publicly available after acceptance in https://github.com/SurgicalDataScienceKCL/NeuralShift.
Enabling High-Curvature Navigation in Eversion Robots through Buckle-Inducing Constrictive Bands
Jan 18, 2026Abstract:Tip-growing eversion robots are renowned for their ability to access remote spaces through narrow passages. However, achieving reliable navigation remains a significant challenge. Existing solutions often rely on artificial muscles integrated into the robot body or active tip-steering mechanisms. While effective, these additions introduce structural complexity and compromise the defining advantages of eversion robots: their inherent softness and compliance. In this paper, we propose a passive approach to reduce bending stiffness by purposefully introducing buckling points along the robot's outer wall. We achieve this by integrating inextensible diameter-reducing circumferential bands at regular intervals along the robot body facilitating forward motion through tortuous, obstacle cluttered paths. Rather than relying on active steering, our approach leverages the robot's natural interaction with the environment, allowing for smooth, compliant navigation. We present a Cosserat rod-based mathematical model to quantify this behavior, capturing the local stiffness reductions caused by the constricting bands and their impact on global bending mechanics. Experimental results demonstrate that these bands reduce the robot's stiffness when bent at the tip by up to 91 percent, enabling consistent traversal of 180 degree bends with a bending radius of as low as 25 mm-notably lower than the 35 mm achievable by standard eversion robots under identical conditions. The feasibility of the proposed method is further demonstrated through a case study in a colon phantom. By significantly improving maneuverability without sacrificing softness or increasing mechanical complexity, this approach expands the applicability of eversion robots in highly curved pathways, whether in relation to pipe inspection or medical procedures such as colonoscopy.
DIST-CLIP: Arbitrary Metadata and Image Guided MRI Harmonization via Disentangled Anatomy-Contrast Representations
Dec 08, 2025Abstract:Deep learning holds immense promise for transforming medical image analysis, yet its clinical generalization remains profoundly limited. A major barrier is data heterogeneity. This is particularly true in Magnetic Resonance Imaging, where scanner hardware differences, diverse acquisition protocols, and varying sequence parameters introduce substantial domain shifts that obscure underlying biological signals. Data harmonization methods aim to reduce these instrumental and acquisition variability, but existing approaches remain insufficient. When applied to imaging data, image-based harmonization approaches are often restricted by the need for target images, while existing text-guided methods rely on simplistic labels that fail to capture complex acquisition details or are typically restricted to datasets with limited variability, failing to capture the heterogeneity of real-world clinical environments. To address these limitations, we propose DIST-CLIP (Disentangled Style Transfer with CLIP Guidance), a unified framework for MRI harmonization that flexibly uses either target images or DICOM metadata for guidance. Our framework explicitly disentangles anatomical content from image contrast, with the contrast representations being extracted using pre-trained CLIP encoders. These contrast embeddings are then integrated into the anatomical content via a novel Adaptive Style Transfer module. We trained and evaluated DIST-CLIP on diverse real-world clinical datasets, and showed significant improvements in performance when compared against state-of-the-art methods in both style translation fidelity and anatomical preservation, offering a flexible solution for style transfer and standardizing MRI data. Our code and weights will be made publicly available upon publication.
LIHE: Linguistic Instance-Split Hyperbolic-Euclidean Framework for Generalized Weakly-Supervised Referring Expression Comprehension
Nov 15, 2025



Abstract:Existing Weakly-Supervised Referring Expression Comprehension (WREC) methods, while effective, are fundamentally limited by a one-to-one mapping assumption, hindering their ability to handle expressions corresponding to zero or multiple targets in realistic scenarios. To bridge this gap, we introduce the Weakly-Supervised Generalized Referring Expression Comprehension task (WGREC), a more practical paradigm that handles expressions with variable numbers of referents. However, extending WREC to WGREC presents two fundamental challenges: supervisory signal ambiguity, where weak image-level supervision is insufficient for training a model to infer the correct number and identity of referents, and semantic representation collapse, where standard Euclidean similarity forces hierarchically-related concepts into non-discriminative clusters, blurring categorical boundaries. To tackle these challenges, we propose a novel WGREC framework named Linguistic Instance-Split Hyperbolic-Euclidean (LIHE), which operates in two stages. The first stage, Referential Decoupling, predicts the number of target objects and decomposes the complex expression into simpler sub-expressions. The second stage, Referent Grounding, then localizes these sub-expressions using HEMix, our innovative hybrid similarity module that synergistically combines the precise alignment capabilities of Euclidean proximity with the hierarchical modeling strengths of hyperbolic geometry. This hybrid approach effectively prevents semantic collapse while preserving fine-grained distinctions between related concepts. Extensive experiments demonstrate LIHE establishes the first effective weakly supervised WGREC baseline on gRefCOCO and Ref-ZOM, while HEMix achieves consistent improvements on standard REC benchmarks, improving IoU@0.5 by up to 2.5\%. The code is available at https://anonymous.4open.science/r/LIHE.
Dynamic causal discovery in Alzheimer's disease through latent pseudotime modelling
Nov 06, 2025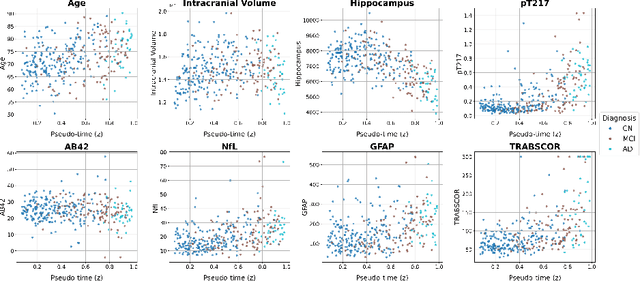



Abstract:The application of causal discovery to diseases like Alzheimer's (AD) is limited by the static graph assumptions of most methods; such models cannot account for an evolving pathophysiology, modulated by a latent disease pseudotime. We propose to apply an existing latent variable model to real-world AD data, inferring a pseudotime that orders patients along a data-driven disease trajectory independent of chronological age, then learning how causal relationships evolve. Pseudotime outperformed age in predicting diagnosis (AUC 0.82 vs 0.59). Incorporating minimal, disease-agnostic background knowledge substantially improved graph accuracy and orientation. Our framework reveals dynamic interactions between novel (NfL, GFAP) and established AD markers, enabling practical causal discovery despite violated assumptions.
A methodology for clinically driven interactive segmentation evaluation
Oct 10, 2025Abstract:Interactive segmentation is a promising strategy for building robust, generalisable algorithms for volumetric medical image segmentation. However, inconsistent and clinically unrealistic evaluation hinders fair comparison and misrepresents real-world performance. We propose a clinically grounded methodology for defining evaluation tasks and metrics, and built a software framework for constructing standardised evaluation pipelines. We evaluate state-of-the-art algorithms across heterogeneous and complex tasks and observe that (i) minimising information loss when processing user interactions is critical for model robustness, (ii) adaptive-zooming mechanisms boost robustness and speed convergence, (iii) performance drops if validation prompting behaviour/budgets differ from training, (iv) 2D methods perform well with slab-like images and coarse targets, but 3D context helps with large or irregularly shaped targets, (v) performance of non-medical-domain models (e.g. SAM2) degrades with poor contrast and complex shapes.
WeakMCN: Multi-task Collaborative Network for Weakly Supervised Referring Expression Comprehension and Segmentation
May 24, 2025Abstract:Weakly supervised referring expression comprehension(WREC) and segmentation(WRES) aim to learn object grounding based on a given expression using weak supervision signals like image-text pairs. While these tasks have traditionally been modeled separately, we argue that they can benefit from joint learning in a multi-task framework. To this end, we propose WeakMCN, a novel multi-task collaborative network that effectively combines WREC and WRES with a dual-branch architecture. Specifically, the WREC branch is formulated as anchor-based contrastive learning, which also acts as a teacher to supervise the WRES branch. In WeakMCN, we propose two innovative designs to facilitate multi-task collaboration, namely Dynamic Visual Feature Enhancement(DVFE) and Collaborative Consistency Module(CCM). DVFE dynamically combines various pre-trained visual knowledge to meet different task requirements, while CCM promotes cross-task consistency from the perspective of optimization. Extensive experimental results on three popular REC and RES benchmarks, i.e., RefCOCO, RefCOCO+, and RefCOCOg, consistently demonstrate performance gains of WeakMCN over state-of-the-art single-task alternatives, e.g., up to 3.91% and 13.11% on RefCOCO for WREC and WRES tasks, respectively. Furthermore, experiments also validate the strong generalization ability of WeakMCN in both semi-supervised REC and RES settings against existing methods, e.g., +8.94% for semi-REC and +7.71% for semi-RES on 1% RefCOCO. The code is publicly available at https://github.com/MRUIL/WeakMCN.
Advancing Embodied Intelligence in Robotic-Assisted Endovascular Procedures: A Systematic Review of AI Solutions
Apr 21, 2025Abstract:Endovascular procedures have revolutionized the treatment of vascular diseases thanks to minimally invasive solutions that significantly reduce patient recovery time and enhance clinical outcomes. However, the precision and dexterity required during these procedures poses considerable challenges for interventionists. Robotic systems have emerged offering transformative solutions, addressing issues such as operator fatigue, radiation exposure, and the inherent limitations of human precision. The integration of Embodied Intelligence (EI) into these systems signifies a paradigm shift, enabling robots to navigate complex vascular networks and adapt to dynamic physiological conditions. Data-driven approaches, advanced computer vision, medical image analysis, and machine learning techniques, are at the forefront of this evolution. These methods augment procedural intelligence by facilitating real-time vessel segmentation, device tracking, and anatomical landmark detection. Reinforcement learning and imitation learning further refine navigation strategies and replicate experts' techniques. This review systematically examines the integration of EI principles into robotic technologies, in relation to endovascular procedures. We discuss recent advancements in intelligent perception and data-driven control, and their practical applications in robot-assisted endovascular procedures. By critically evaluating current limitations and emerging opportunities, this review establishes a framework for future developments, emphasizing the potential for greater autonomy and improved clinical outcomes. Emerging trends and specific areas of research, such as federated learning for medical data sharing, explainable AI for clinical decision support, and advanced human-robot collaboration paradigms, are also explored, offering insights into the future direction of this rapidly evolving field.
UltraFlwr -- An Efficient Federated Medical and Surgical Object Detection Framework
Mar 19, 2025Abstract:Object detection shows promise for medical and surgical applications such as cell counting and tool tracking. However, its faces multiple real-world edge deployment challenges including limited high-quality annotated data, data sharing restrictions, and computational constraints. In this work, we introduce UltraFlwr, a framework for federated medical and surgical object detection. By leveraging Federated Learning (FL), UltraFlwr enables decentralized model training across multiple sites without sharing raw data. To further enhance UltraFlwr's efficiency, we propose YOLO-PA, a set of novel Partial Aggregation (PA) strategies specifically designed for YOLO models in FL. YOLO-PA significantly reduces communication overhead by up to 83% per round while maintaining performance comparable to Full Aggregation (FA) strategies. Our extensive experiments on BCCD and m2cai16-tool-locations datasets demonstrate that YOLO-PA not only provides better client models compared to client-wise centralized training and FA strategies, but also facilitates efficient training and deployment across resource-constrained edge devices. Further, we also establish one of the first benchmarks in federated medical and surgical object detection. This paper advances the feasibility of training and deploying detection models on the edge, making federated object detection more practical for time-critical and resource-constrained medical and surgical applications. UltraFlwr is publicly available at https://github.com/KCL-BMEIS/UltraFlwr.
Resolution Invariant Autoencoder
Mar 12, 2025Abstract:Deep learning has significantly advanced medical imaging analysis, yet variations in image resolution remain an overlooked challenge. Most methods address this by resampling images, leading to either information loss or computational inefficiencies. While solutions exist for specific tasks, no unified approach has been proposed. We introduce a resolution-invariant autoencoder that adapts spatial resizing at each layer in the network via a learned variable resizing process, replacing fixed spatial down/upsampling at the traditional factor of 2. This ensures a consistent latent space resolution, regardless of input or output resolution. Our model enables various downstream tasks to be performed on an image latent whilst maintaining performance across different resolutions, overcoming the shortfalls of traditional methods. We demonstrate its effectiveness in uncertainty-aware super-resolution, classification, and generative modelling tasks and show how our method outperforms conventional baselines with minimal performance loss across resolutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge