Tobias Roß
Labeling instructions matter in biomedical image analysis
Jul 20, 2022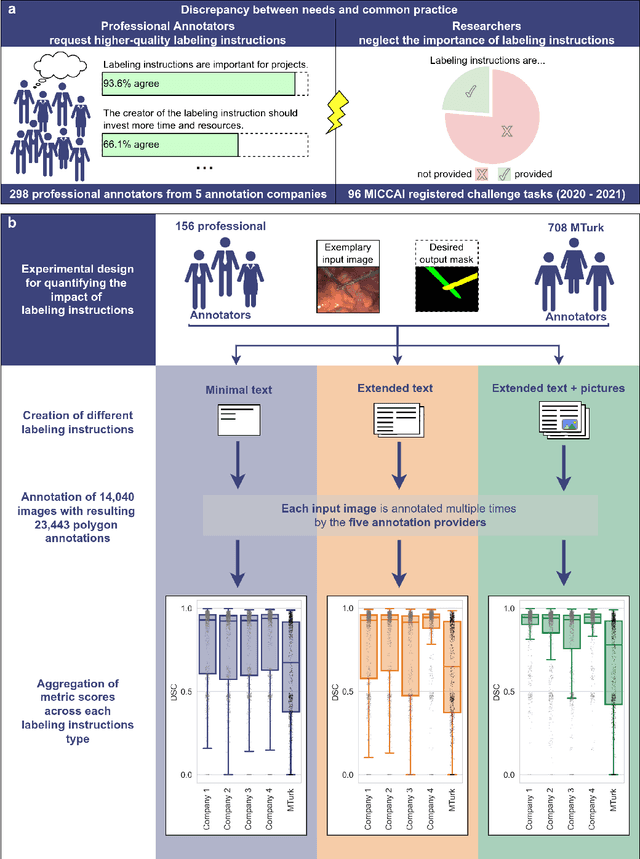
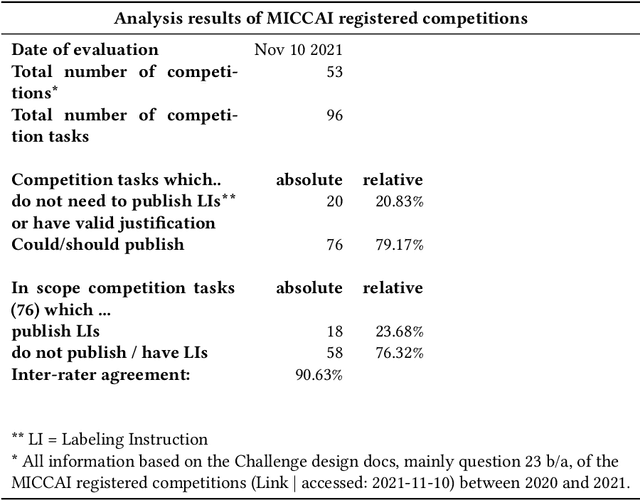
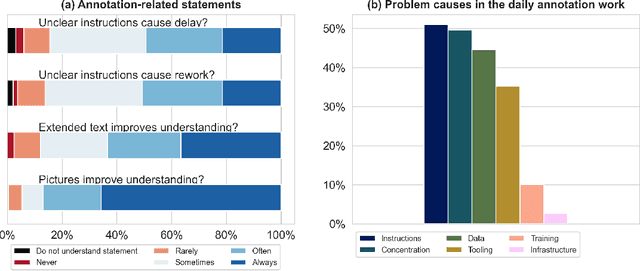
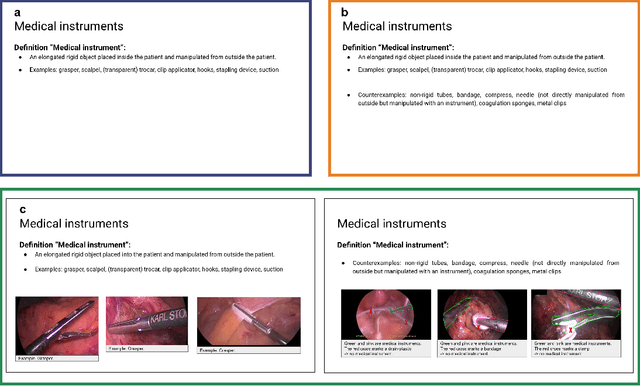
Abstract:Biomedical image analysis algorithm validation depends on high-quality annotation of reference datasets, for which labeling instructions are key. Despite their importance, their optimization remains largely unexplored. Here, we present the first systematic study of labeling instructions and their impact on annotation quality in the field. Through comprehensive examination of professional practice and international competitions registered at the MICCAI Society, we uncovered a discrepancy between annotators' needs for labeling instructions and their current quality and availability. Based on an analysis of 14,040 images annotated by 156 annotators from four professional companies and 708 Amazon Mechanical Turk (MTurk) crowdworkers using instructions with different information density levels, we further found that including exemplary images significantly boosts annotation performance compared to text-only descriptions, while solely extending text descriptions does not. Finally, professional annotators constantly outperform MTurk crowdworkers. Our study raises awareness for the need of quality standards in biomedical image analysis labeling instructions.
How can we learn from challenges? A statistical approach to driving future algorithm development
Jun 17, 2021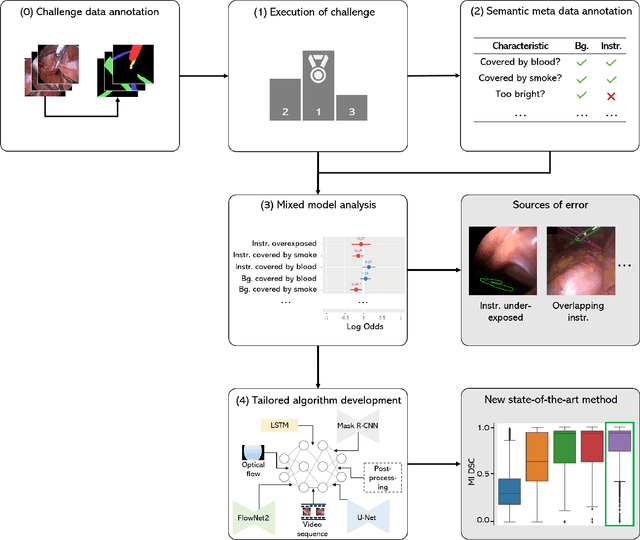
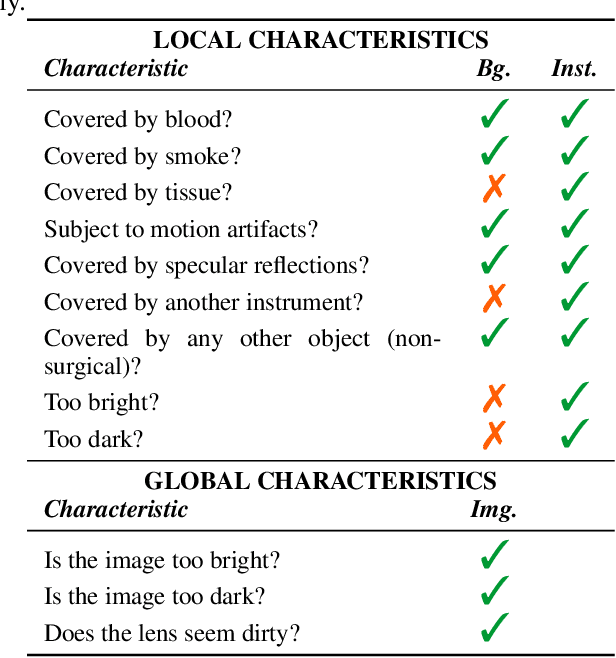
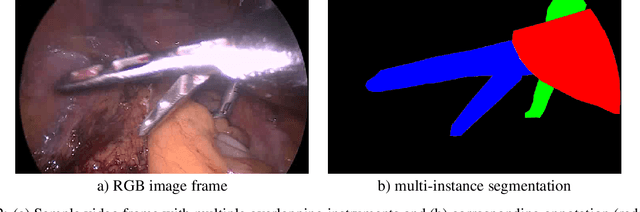
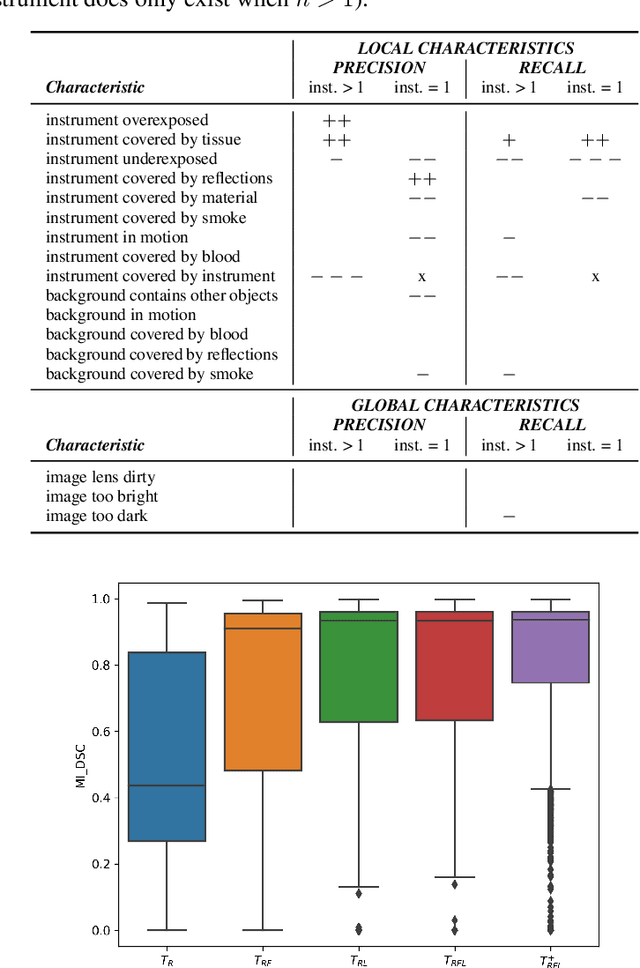
Abstract:Challenges have become the state-of-the-art approach to benchmark image analysis algorithms in a comparative manner. While the validation on identical data sets was a great step forward, results analysis is often restricted to pure ranking tables, leaving relevant questions unanswered. Specifically, little effort has been put into the systematic investigation on what characterizes images in which state-of-the-art algorithms fail. To address this gap in the literature, we (1) present a statistical framework for learning from challenges and (2) instantiate it for the specific task of instrument instance segmentation in laparoscopic videos. Our framework relies on the semantic meta data annotation of images, which serves as foundation for a General Linear Mixed Models (GLMM) analysis. Based on 51,542 meta data annotations performed on 2,728 images, we applied our approach to the results of the Robust Medical Instrument Segmentation Challenge (ROBUST-MIS) challenge 2019 and revealed underexposure, motion and occlusion of instruments as well as the presence of smoke or other objects in the background as major sources of algorithm failure. Our subsequent method development, tailored to the specific remaining issues, yielded a deep learning model with state-of-the-art overall performance and specific strengths in the processing of images in which previous methods tended to fail. Due to the objectivity and generic applicability of our approach, it could become a valuable tool for validation in the field of medical image analysis and beyond. and segmentation of small, crossing, moving and transparent instrument(s) (parts).
Generating large labeled data sets for laparoscopic image processing tasks using unpaired image-to-image translation
Jul 05, 2019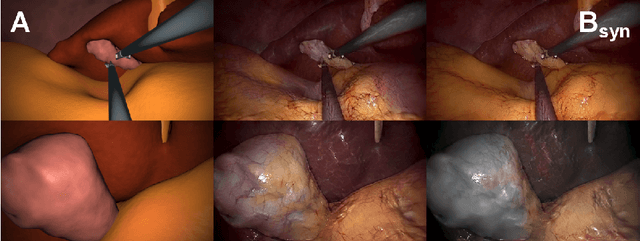
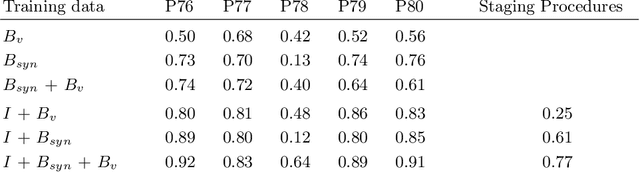
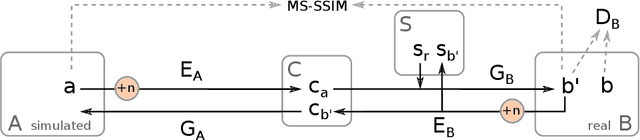
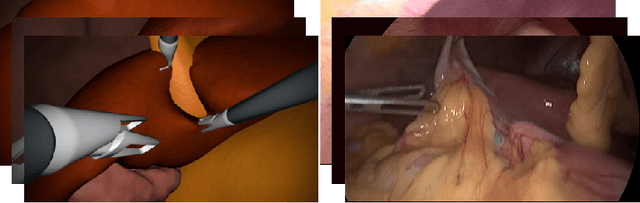
Abstract:In the medical domain, the lack of large training data sets and benchmarks is often a limiting factor for training deep neural networks. In contrast to expensive manual labeling, computer simulations can generate large and fully labeled data sets with a minimum of manual effort. However, models that are trained on simulated data usually do not translate well to real scenarios. To bridge the domain gap between simulated and real laparoscopic images, we exploit recent advances in unpaired image-to-image translation. We extent an image-to-image translation method to generate a diverse multitude of realistically looking synthetic images based on images from a simple laparoscopy simulation. By incorporating means to ensure that the image content is preserved during the translation process, we ensure that the labels given for the simulated images remain valid for their realistically looking translations. This way, we are able to generate a large, fully labeled synthetic data set of laparoscopic images with realistic appearance. We show that this data set can be used to train models for the task of liver segmentation of laparoscopic images. We achieve average dice scores of up to 0.89 in some patients without manually labeling a single laparoscopic image and show that using our synthetic data to pre-train models can greatly improve their performance. The synthetic data set will be made publicly available, fully labeled with segmentation maps, depth maps, normal maps, and positions of tools and camera (http://opencas.dkfz.de/image2image).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge