Gaël Varoquaux
SODA
Scalable Feature Learning on Huge Knowledge Graphs for Downstream Machine Learning
Jul 01, 2025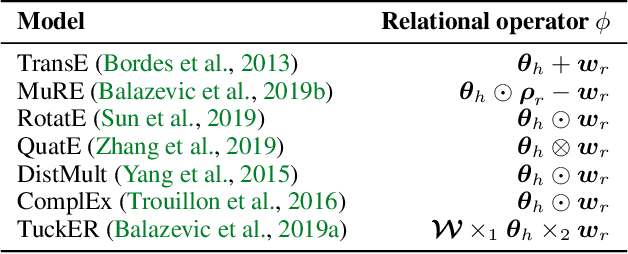
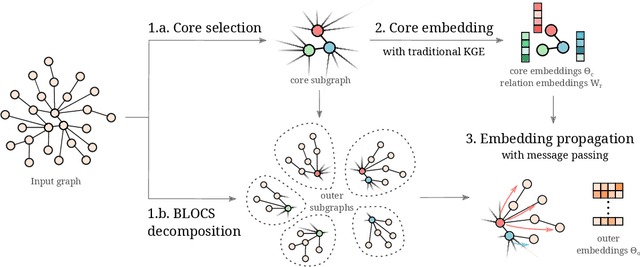
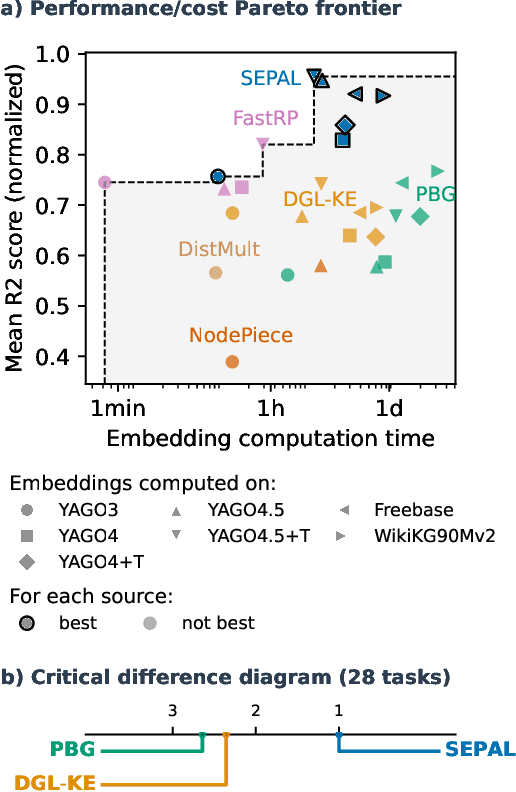
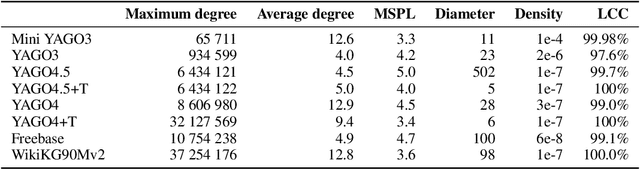
Abstract:Many machine learning tasks can benefit from external knowledge. Large knowledge graphs store such knowledge, and embedding methods can be used to distill it into ready-to-use vector representations for downstream applications. For this purpose, current models have however two limitations: they are primarily optimized for link prediction, via local contrastive learning, and they struggle to scale to the largest graphs due to GPU memory limits. To address these, we introduce SEPAL: a Scalable Embedding Propagation ALgorithm for large knowledge graphs designed to produce high-quality embeddings for downstream tasks at scale. The key idea of SEPAL is to enforce global embedding alignment by optimizing embeddings only on a small core of entities, and then propagating them to the rest of the graph via message passing. We evaluate SEPAL on 7 large-scale knowledge graphs and 46 downstream machine learning tasks. Our results show that SEPAL significantly outperforms previous methods on downstream tasks. In addition, SEPAL scales up its base embedding model, enabling fitting huge knowledge graphs on commodity hardware.
Query-Level Uncertainty in Large Language Models
Jun 11, 2025Abstract:It is important for Large Language Models to be aware of the boundary of their knowledge, the mechanism of identifying known and unknown queries. This type of awareness can help models perform adaptive inference, such as invoking RAG, engaging in slow and deep thinking, or adopting the abstention mechanism, which is beneficial to the development of efficient and trustworthy AI. In this work, we propose a method to detect knowledge boundaries via Query-Level Uncertainty, which aims to determine if the model is able to address a given query without generating any tokens. To this end, we introduce a novel and training-free method called \emph{Internal Confidence}, which leverages self-evaluations across layers and tokens. Empirical results on both factual QA and mathematical reasoning tasks demonstrate that our internal confidence can outperform several baselines. Furthermore, we showcase that our proposed method can be used for efficient RAG and model cascading, which is able to reduce inference costs while maintaining performance.
Table Foundation Models: on knowledge pre-training for tabular learning
May 20, 2025Abstract:Table foundation models bring high hopes to data science: pre-trained on tabular data to embark knowledge or priors, they should facilitate downstream tasks on tables. One specific challenge is that of data semantics: numerical entries take their meaning from context, e.g., column name. Pre-trained neural networks that jointly model column names and table entries have recently boosted prediction accuracy. While these models outline the promises of world knowledge to interpret table values, they lack the convenience of popular foundation models in text or vision. Indeed, they must be fine-tuned to bring benefits, come with sizeable computation costs, and cannot easily be reused or combined with other architectures. Here we introduce TARTE, a foundation model that transforms tables to knowledge-enhanced vector representations using the string to capture semantics. Pre-trained on large relational data, TARTE yields representations that facilitate subsequent learning with little additional cost. These representations can be fine-tuned or combined with other learners, giving models that push the state-of-the-art prediction performance and improve the prediction/computation performance trade-off. Specialized to a task or a domain, TARTE gives domain-specific representations that facilitate further learning. Our study demonstrates an effective approach to knowledge pre-training for tabular learning.
False Promises in Medical Imaging AI? Assessing Validity of Outperformance Claims
May 07, 2025Abstract:Performance comparisons are fundamental in medical imaging Artificial Intelligence (AI) research, often driving claims of superiority based on relative improvements in common performance metrics. However, such claims frequently rely solely on empirical mean performance. In this paper, we investigate whether newly proposed methods genuinely outperform the state of the art by analyzing a representative cohort of medical imaging papers. We quantify the probability of false claims based on a Bayesian approach that leverages reported results alongside empirically estimated model congruence to estimate whether the relative ranking of methods is likely to have occurred by chance. According to our results, the majority (>80%) of papers claims outperformance when introducing a new method. Our analysis further revealed a high probability (>5%) of false outperformance claims in 86% of classification papers and 53% of segmentation papers. These findings highlight a critical flaw in current benchmarking practices: claims of outperformance in medical imaging AI are frequently unsubstantiated, posing a risk of misdirecting future research efforts.
TabICL: A Tabular Foundation Model for In-Context Learning on Large Data
Feb 08, 2025



Abstract:The long-standing dominance of gradient-boosted decision trees on tabular data is currently challenged by tabular foundation models using In-Context Learning (ICL): setting the training data as context for the test data and predicting in a single forward pass without parameter updates. While the very recent TabPFNv2 foundation model (2025) excels on tables with up to 10K samples, its alternating column- and row-wise attentions make handling large training sets computationally prohibitive. So, can ICL be effectively scaled and deliver a benefit for larger tables? We introduce TabICL, a tabular foundation model for classification, pretrained on synthetic datasets with up to 60K samples and capable of handling 500K samples on affordable resources. This is enabled by a novel two-stage architecture: a column-then-row attention mechanism to build fixed-dimensional embeddings of rows, followed by a transformer for efficient ICL. Across 200 classification datasets from the TALENT benchmark, TabICL is on par with TabPFNv2 while being systematically faster (up to 10 times), and significantly outperforms all other approaches. On 56 datasets with over 10K samples, TabICL surpasses both TabPFNv2 and CatBoost, demonstrating the potential of ICL for large data.
International AI Safety Report
Jan 29, 2025



Abstract:The first International AI Safety Report comprehensively synthesizes the current evidence on the capabilities, risks, and safety of advanced AI systems. The report was mandated by the nations attending the AI Safety Summit in Bletchley, UK. Thirty nations, the UN, the OECD, and the EU each nominated a representative to the report's Expert Advisory Panel. A total of 100 AI experts contributed, representing diverse perspectives and disciplines. Led by the report's Chair, these independent experts collectively had full discretion over the report's content.
Survival Models: Proper Scoring Rule and Stochastic Optimization with Competing Risks
Oct 22, 2024Abstract:When dealing with right-censored data, where some outcomes are missing due to a limited observation period, survival analysis -- known as time-to-event analysis -- focuses on predicting the time until an event of interest occurs. Multiple classes of outcomes lead to a classification variant: predicting the most likely event, a less explored area known as competing risks. Classic competing risks models couple architecture and loss, limiting scalability.To address these issues, we design a strictly proper censoring-adjusted separable scoring rule, allowing optimization on a subset of the data as each observation is evaluated independently. The loss estimates outcome probabilities and enables stochastic optimization for competing risks, which we use for efficient gradient boosting trees. SurvivalBoost not only outperforms 12 state-of-the-art models across several metrics on 4 real-life datasets, both in competing risks and survival settings, but also provides great calibration, the ability to predict across any time horizon, and computation times faster than existing methods.
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024



Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
What is the Role of Small Models in the LLM Era: A Survey
Sep 12, 2024



Abstract:Large Language Models (LLMs) have made significant progress in advancing artificial general intelligence (AGI), leading to the development of increasingly large models such as GPT-4 and LLaMA-405B. However, scaling up model sizes results in exponentially higher computational costs and energy consumption, making these models impractical for academic researchers and businesses with limited resources. At the same time, Small Models (SMs) are frequently used in practical settings, although their significance is currently underestimated. This raises important questions about the role of small models in the era of LLMs, a topic that has received limited attention in prior research. In this work, we systematically examine the relationship between LLMs and SMs from two key perspectives: Collaboration and Competition. We hope this survey provides valuable insights for practitioners, fostering a deeper understanding of the contribution of small models and promoting more efficient use of computational resources. The code is available at https://github.com/tigerchen52/role_of_small_models
Imputation for prediction: beware of diminishing returns
Jul 29, 2024



Abstract:Missing values are prevalent across various fields, posing challenges for training and deploying predictive models. In this context, imputation is a common practice, driven by the hope that accurate imputations will enhance predictions. However, recent theoretical and empirical studies indicate that simple constant imputation can be consistent and competitive. This empirical study aims at clarifying if and when investing in advanced imputation methods yields significantly better predictions. Relating imputation and predictive accuracies across combinations of imputation and predictive models on 20 datasets, we show that imputation accuracy matters less i) when using expressive models, ii) when incorporating missingness indicators as complementary inputs, iii) matters much more for generated linear outcomes than for real-data outcomes. Interestingly, we also show that the use of the missingness indicator is beneficial to the prediction performance, even in MCAR scenarios. Overall, on real-data with powerful models, improving imputation only has a minor effect on prediction performance. Thus, investing in better imputations for improved predictions often offers limited benefits.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge