Debesh Jha
From SAM to DINOv2: Towards Distilling Foundation Models to Lightweight Baselines for Generalized Polyp Segmentation
Dec 10, 2025Abstract:Accurate polyp segmentation during colonoscopy is critical for the early detection of colorectal cancer and still remains challenging due to significant size, shape, and color variations, and the camouflaged nature of polyps. While lightweight baseline models such as U-Net, U-Net++, and PraNet offer advantages in terms of easy deployment and low computational cost, they struggle to deal with the above issues, leading to limited segmentation performance. In contrast, large-scale vision foundation models such as SAM, DINOv2, OneFormer, and Mask2Former have exhibited impressive generalization performance across natural image domains. However, their direct transfer to medical imaging tasks (e.g., colonoscopic polyp segmentation) is not straightforward, primarily due to the scarcity of large-scale datasets and lack of domain-specific knowledge. To bridge this gap, we propose a novel distillation framework, Polyp-DiFoM, that transfers the rich representations of foundation models into lightweight segmentation baselines, allowing efficient and accurate deployment in clinical settings. In particular, we infuse semantic priors from the foundation models into canonical architectures such as U-Net and U-Net++ and further perform frequency domain encoding for enhanced distillation, corroborating their generalization capability. Extensive experiments are performed across five benchmark datasets, such as Kvasir-SEG, CVC-ClinicDB, ETIS, ColonDB, and CVC-300. Notably, Polyp-DiFoM consistently outperforms respective baseline models significantly, as well as the state-of-the-art model, with nearly 9 times reduced computation overhead. The code is available at https://github.com/lostinrepo/PolypDiFoM.
When CNNs Outperform Transformers and Mambas: Revisiting Deep Architectures for Dental Caries Segmentation
Nov 18, 2025



Abstract:Accurate identification and segmentation of dental caries in panoramic radiographs are critical for early diagnosis and effective treatment planning. Automated segmentation remains challenging due to low lesion contrast, morphological variability, and limited annotated data. In this study, we present the first comprehensive benchmarking of convolutional neural networks, vision transformers and state-space mamba architectures for automated dental caries segmentation on panoramic radiographs through a DC1000 dataset. Twelve state-of-the-art architectures, including VMUnet, MambaUNet, VMUNetv2, RMAMamba-S, TransNetR, PVTFormer, DoubleU-Net, and ResUNet++, were trained under identical configurations. Results reveal that, contrary to the growing trend toward complex attention based architectures, the CNN-based DoubleU-Net achieved the highest dice coefficient of 0.7345, mIoU of 0.5978, and precision of 0.8145, outperforming all transformer and Mamba variants. In the study, the top 3 results across all performance metrics were achieved by CNN-based architectures. Here, Mamba and transformer-based methods, despite their theoretical advantage in global context modeling, underperformed due to limited data and weaker spatial priors. These findings underscore the importance of architecture-task alignment in domain-specific medical image segmentation more than model complexity. Our code is available at: https://github.com/JunZengz/dental-caries-segmentation.
Viper-F1: Fast and Fine-Grained Multimodal Understanding with Cross-Modal State-Space Modulation
Nov 18, 2025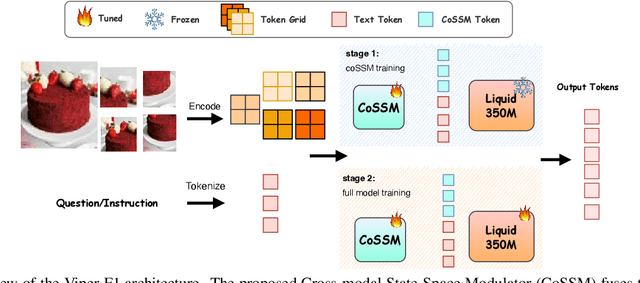
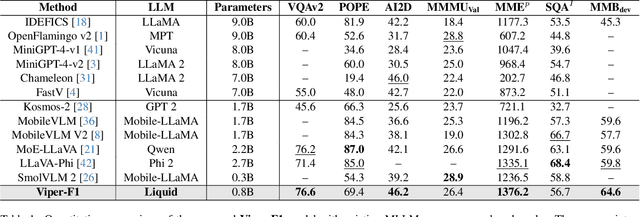
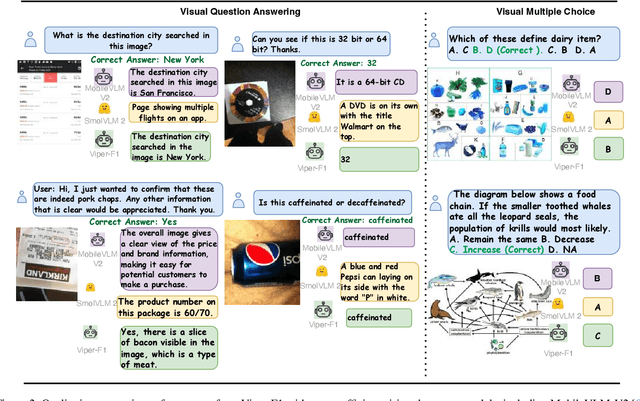
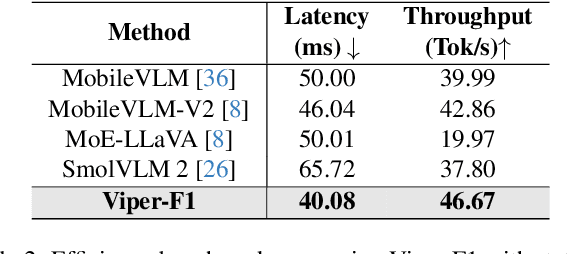
Abstract:Recent advances in multimodal large language models (MLLMs) have enabled impressive progress in vision-language understanding, yet their high computational cost limits deployment in resource-constrained scenarios such as robotic manipulation, personal assistants, and smart cameras. Most existing methods rely on Transformer-based cross-attention, whose quadratic complexity hinders efficiency. Moreover, small vision-language models often struggle to precisely capture fine-grained, task-relevant visual regions, leading to degraded performance on fine-grained reasoning tasks that limit their effectiveness in the real world. To address these issues, we introduce Viper-F1, a Hybrid State-Space Vision-Language Model that replaces attention with efficient Liquid State-Space Dynamics. To further enhance visual grounding, we propose a Token-Grid Correlation Module, which computes lightweight correlations between text tokens and image patches and modulates the state-space dynamics via FiLM conditioning. This enables the model to selectively emphasize visual regions relevant to the textual prompt while maintaining linear-time inference. Experimental results across multiple benchmarks demonstrate that Viper-F1 achieves accurate, fine-grained understanding with significantly improved efficiency.
SRMA-Mamba: Spatial Reverse Mamba Attention Network for Pathological Liver Segmentation in MRI Volumes
Aug 17, 2025



Abstract:Liver Cirrhosis plays a critical role in the prognosis of chronic liver disease. Early detection and timely intervention are critical in significantly reducing mortality rates. However, the intricate anatomical architecture and diverse pathological changes of liver tissue complicate the accurate detection and characterization of lesions in clinical settings. Existing methods underutilize the spatial anatomical details in volumetric MRI data, thereby hindering their clinical effectiveness and explainability. To address this challenge, we introduce a novel Mamba-based network, SRMA-Mamba, designed to model the spatial relationships within the complex anatomical structures of MRI volumes. By integrating the Spatial Anatomy-Based Mamba module (SABMamba), SRMA-Mamba performs selective Mamba scans within liver cirrhotic tissues and combines anatomical information from the sagittal, coronal, and axial planes to construct a global spatial context representation, enabling efficient volumetric segmentation of pathological liver structures. Furthermore, we introduce the Spatial Reverse Attention module (SRMA), designed to progressively refine cirrhotic details in the segmentation map, utilizing both the coarse segmentation map and hierarchical encoding features. Extensive experiments demonstrate that SRMA-Mamba surpasses state-of-the-art methods, delivering exceptional performance in 3D pathological liver segmentation. Our code is available for public: {\color{blue}{https://github.com/JunZengz/SRMA-Mamba}}.
Large Language Model Evaluated Stand-alone Attention-Assisted Graph Neural Network with Spatial and Structural Information Interaction for Precise Endoscopic Image Segmentation
Aug 09, 2025Abstract:Accurate endoscopic image segmentation on the polyps is critical for early colorectal cancer detection. However, this task remains challenging due to low contrast with surrounding mucosa, specular highlights, and indistinct boundaries. To address these challenges, we propose FOCUS-Med, which stands for Fusion of spatial and structural graph with attentional context-aware polyp segmentation in endoscopic medical imaging. FOCUS-Med integrates a Dual Graph Convolutional Network (Dual-GCN) module to capture contextual spatial and topological structural dependencies. This graph-based representation enables the model to better distinguish polyps from background tissues by leveraging topological cues and spatial connectivity, which are often obscured in raw image intensities. It enhances the model's ability to preserve boundaries and delineate complex shapes typical of polyps. In addition, a location-fused stand-alone self-attention is employed to strengthen global context integration. To bridge the semantic gap between encoder-decoder layers, we incorporate a trainable weighted fast normalized fusion strategy for efficient multi-scale aggregation. Notably, we are the first to introduce the use of a Large Language Model (LLM) to provide detailed qualitative evaluations of segmentation quality. Extensive experiments on public benchmarks demonstrate that FOCUS-Med achieves state-of-the-art performance across five key metrics, underscoring its effectiveness and clinical potential for AI-assisted colonoscopy.
Mamba Guided Boundary Prior Matters: A New Perspective for Generalized Polyp Segmentation
Jul 02, 2025Abstract:Polyp segmentation in colonoscopy images is crucial for early detection and diagnosis of colorectal cancer. However, this task remains a significant challenge due to the substantial variations in polyp shape, size, and color, as well as the high similarity between polyps and surrounding tissues, often compounded by indistinct boundaries. While existing encoder-decoder CNN and transformer-based approaches have shown promising results, they struggle with stable segmentation performance on polyps with weak or blurry boundaries. These methods exhibit limited abilities to distinguish between polyps and non-polyps and capture essential boundary cues. Moreover, their generalizability still falls short of meeting the demands of real-time clinical applications. To address these limitations, we propose SAM-MaGuP, a groundbreaking approach for robust polyp segmentation. By incorporating a boundary distillation module and a 1D-2D Mamba adapter within the Segment Anything Model (SAM), SAM-MaGuP excels at resolving weak boundary challenges and amplifies feature learning through enriched global contextual interactions. Extensive evaluations across five diverse datasets reveal that SAM-MaGuP outperforms state-of-the-art methods, achieving unmatched segmentation accuracy and robustness. Our key innovations, a Mamba-guided boundary prior and a 1D-2D Mamba block, set a new benchmark in the field, pushing the boundaries of polyp segmentation to new heights.
PRS-Med: Position Reasoning Segmentation with Vision-Language Model in Medical Imaging
May 17, 2025



Abstract:Recent advancements in prompt-based medical image segmentation have enabled clinicians to identify tumors using simple input like bounding boxes or text prompts. However, existing methods face challenges when doctors need to interact through natural language or when position reasoning is required - understanding spatial relationships between anatomical structures and pathologies. We present PRS-Med, a framework that integrates vision-language models with segmentation capabilities to generate both accurate segmentation masks and corresponding spatial reasoning outputs. Additionally, we introduce the MMRS dataset (Multimodal Medical in Positional Reasoning Segmentation), which provides diverse, spatially-grounded question-answer pairs to address the lack of position reasoning data in medical imaging. PRS-Med demonstrates superior performance across six imaging modalities (CT, MRI, X-ray, ultrasound, endoscopy, RGB), significantly outperforming state-of-the-art methods in both segmentation accuracy and position reasoning. Our approach enables intuitive doctor-system interaction through natural language, facilitating more efficient diagnoses. Our dataset pipeline, model, and codebase will be released to foster further research in spatially-aware multimodal reasoning for medical applications.
FocusNet: Transformer-enhanced Polyp Segmentation with Local and Pooling Attention
Apr 18, 2025Abstract:Colonoscopy is vital in the early diagnosis of colorectal polyps. Regular screenings can effectively prevent benign polyps from progressing to CRC. While deep learning has made impressive strides in polyp segmentation, most existing models are trained on single-modality and single-center data, making them less effective in real-world clinical environments. To overcome these limitations, we propose FocusNet, a Transformer-enhanced focus attention network designed to improve polyp segmentation. FocusNet incorporates three essential modules: the Cross-semantic Interaction Decoder Module (CIDM) for generating coarse segmentation maps, the Detail Enhancement Module (DEM) for refining shallow features, and the Focus Attention Module (FAM), to balance local detail and global context through local and pooling attention mechanisms. We evaluate our model on PolypDB, a newly introduced dataset with multi-modality and multi-center data for building more reliable segmentation methods. Extensive experiments showed that FocusNet consistently outperforms existing state-of-the-art approaches with a high dice coefficients of 82.47% on the BLI modality, 88.46% on FICE, 92.04% on LCI, 82.09% on the NBI and 93.42% on WLI modality, demonstrating its accuracy and robustness across five different modalities. The source code for FocusNet is available at https://github.com/JunZengz/FocusNet.
Liver Cirrhosis Stage Estimation from MRI with Deep Learning
Feb 23, 2025Abstract:We present an end-to-end deep learning framework for automated liver cirrhosis stage estimation from multi-sequence MRI. Cirrhosis is the severe scarring (fibrosis) of the liver and a common endpoint of various chronic liver diseases. Early diagnosis is vital to prevent complications such as decompensation and cancer, which significantly decreases life expectancy. However, diagnosing cirrhosis in its early stages is challenging, and patients often present with life-threatening complications. Our approach integrates multi-scale feature learning with sequence-specific attention mechanisms to capture subtle tissue variations across cirrhosis progression stages. Using CirrMRI600+, a large-scale publicly available dataset of 628 high-resolution MRI scans from 339 patients, we demonstrate state-of-the-art performance in three-stage cirrhosis classification. Our best model achieves 72.8% accuracy on T1W and 63.8% on T2W sequences, significantly outperforming traditional radiomics-based approaches. Through extensive ablation studies, we show that our architecture effectively learns stage-specific imaging biomarkers. We establish new benchmarks for automated cirrhosis staging and provide insights for developing clinically applicable deep learning systems. The source code will be available at https://github.com/JunZengz/CirrhosisStage.
A Reverse Mamba Attention Network for Pathological Liver Segmentation
Feb 23, 2025



Abstract:We present RMA-Mamba, a novel architecture that advances the capabilities of vision state space models through a specialized reverse mamba attention module (RMA). The key innovation lies in RMA-Mamba's ability to capture long-range dependencies while maintaining precise local feature representation through its hierarchical processing pipeline. By integrating Vision Mamba (VMamba)'s efficient sequence modeling with RMA's targeted feature refinement, our architecture achieves superior feature learning across multiple scales. This dual-mechanism approach enables robust handling of complex morphological patterns while maintaining computational efficiency. We demonstrate RMA-Mamba's effectiveness in the challenging domain of pathological liver segmentation (from both CT and MRI), where traditional segmentation approaches often fail due to tissue variations. When evaluated on a newly introduced cirrhotic liver dataset (CirrMRI600+) of T2-weighted MRI scans, RMA-Mamba achieves the state-of-the-art performance with a Dice coefficient of 92.08%, mean IoU of 87.36%, and recall of 92.96%. The architecture's generalizability is further validated on the cancerous liver segmentation from CT scans (LiTS: Liver Tumor Segmentation dataset), yielding a Dice score of 92.9% and mIoU of 88.99%. The source code of the proposed RMA-Mamba is available at https://github.com/JunZengz/RMAMamba.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge