Klaus Maier-Hein
*: shared first/last authors
Finally Outshining the Random Baseline: A Simple and Effective Solution for Active Learning in 3D Biomedical Imaging
Jan 20, 2026Abstract:Active learning (AL) has the potential to drastically reduce annotation costs in 3D biomedical image segmentation, where expert labeling of volumetric data is both time-consuming and expensive. Yet, existing AL methods are unable to consistently outperform improved random sampling baselines adapted to 3D data, leaving the field without a reliable solution. We introduce Class-stratified Scheduled Power Predictive Entropy (ClaSP PE), a simple and effective query strategy that addresses two key limitations of standard uncertainty-based AL methods: class imbalance and redundancy in early selections. ClaSP PE combines class-stratified querying to ensure coverage of underrepresented structures and log-scale power noising with a decaying schedule to enforce query diversity in early-stage AL and encourage exploitation later. In our evaluation on 24 experimental settings using four 3D biomedical datasets within the comprehensive nnActive benchmark, ClaSP PE is the only method that generally outperforms improved random baselines in terms of both segmentation quality with statistically significant gains, whilst remaining annotation efficient. Furthermore, we explicitly simulate the real-world application by testing our method on four previously unseen datasets without manual adaptation, where all experiment parameters are set according to predefined guidelines. The results confirm that ClaSP PE robustly generalizes to novel tasks without requiring dataset-specific tuning. Within the nnActive framework, we present compelling evidence that an AL method can consistently outperform random baselines adapted to 3D segmentation, in terms of both performance and annotation efficiency in a realistic, close-to-production scenario. Our open-source implementation and clear deployment guidelines make it readily applicable in practice. Code is at https://github.com/MIC-DKFZ/nnActive.
MedNeXt-v2: Scaling 3D ConvNeXts for Large-Scale Supervised Representation Learning in Medical Image Segmentation
Dec 19, 2025Abstract:Large-scale supervised pretraining is rapidly reshaping 3D medical image segmentation. However, existing efforts focus primarily on increasing dataset size and overlook the question of whether the backbone network is an effective representation learner at scale. In this work, we address this gap by revisiting ConvNeXt-based architectures for volumetric segmentation and introducing MedNeXt-v2, a compound-scaled 3D ConvNeXt that leverages improved micro-architecture and data scaling to deliver state-of-the-art performance. First, we show that routinely used backbones in large-scale pretraining pipelines are often suboptimal. Subsequently, we use comprehensive backbone benchmarking prior to scaling and demonstrate that stronger from scratch performance reliably predicts stronger downstream performance after pretraining. Guided by these findings, we incorporate a 3D Global Response Normalization module and use depth, width, and context scaling to improve our architecture for effective representation learning. We pretrain MedNeXt-v2 on 18k CT volumes and demonstrate state-of-the-art performance when fine-tuning across six challenging CT and MR benchmarks (144 structures), showing consistent gains over seven publicly released pretrained models. Beyond improvements, our benchmarking of these models also reveals that stronger backbones yield better results on similar data, representation scaling disproportionately benefits pathological segmentation, and that modality-specific pretraining offers negligible benefit once full finetuning is applied. In conclusion, our results establish MedNeXt-v2 as a strong backbone for large-scale supervised representation learning in 3D Medical Image Segmentation. Our code and pretrained models are made available with the official nnUNet repository at: https://www.github.com/MIC-DKFZ/nnUNet
CRONOS: Continuous Time Reconstruction for 4D Medical Longitudinal Series
Dec 18, 2025Abstract:Forecasting how 3D medical scans evolve over time is important for disease progression, treatment planning, and developmental assessment. Yet existing models either rely on a single prior scan, fixed grid times, or target global labels, which limits voxel-level forecasting under irregular sampling. We present CRONOS, a unified framework for many-to-one prediction from multiple past scans that supports both discrete (grid-based) and continuous (real-valued) timestamps in one model, to the best of our knowledge the first to achieve continuous sequence-to-image forecasting for 3D medical data. CRONOS learns a spatio-temporal velocity field that transports context volumes toward a target volume at an arbitrary time, while operating directly in 3D voxel space. Across three public datasets spanning Cine-MRI, perfusion CT, and longitudinal MRI, CRONOS outperforms other baselines, while remaining computationally competitive. We will release code and evaluation protocols to enable reproducible, multi-dataset benchmarking of multi-context, continuous-time forecasting.
Kaapana: A Comprehensive Open-Source Platform for Integrating AI in Medical Imaging Research Environments
Dec 10, 2025
Abstract:Developing generalizable AI for medical imaging requires both access to large, multi-center datasets and standardized, reproducible tooling within research environments. However, leveraging real-world imaging data in clinical research environments is still hampered by strict regulatory constraints, fragmented software infrastructure, and the challenges inherent in conducting large-cohort multicentre studies. This leads to projects that rely on ad-hoc toolchains that are hard to reproduce, difficult to scale beyond single institutions and poorly suited for collaboration between clinicians and data scientists. We present Kaapana, a comprehensive open-source platform for medical imaging research that is designed to bridge this gap. Rather than building single-use, site-specific tooling, Kaapana provides a modular, extensible framework that unifies data ingestion, cohort curation, processing workflows and result inspection under a common user interface. By bringing the algorithm to the data, it enables institutions to keep control over their sensitive data while still participating in distributed experimentation and model development. By integrating flexible workflow orchestration with user-facing applications for researchers, Kaapana reduces technical overhead, improves reproducibility and enables conducting large-scale, collaborative, multi-centre imaging studies. We describe the core concepts of the platform and illustrate how they can support diverse use cases, from local prototyping to nation-wide research networks. The open-source codebase is available at https://github.com/kaapana/kaapana
VoxTell: Free-Text Promptable Universal 3D Medical Image Segmentation
Nov 14, 2025Abstract:We introduce VoxTell, a vision-language model for text-prompted volumetric medical image segmentation. It maps free-form descriptions, from single words to full clinical sentences, to 3D masks. Trained on 62K+ CT, MRI, and PET volumes spanning over 1K anatomical and pathological classes, VoxTell uses multi-stage vision-language fusion across decoder layers to align textual and visual features at multiple scales. It achieves state-of-the-art zero-shot performance across modalities on unseen datasets, excelling on familiar concepts while generalizing to related unseen classes. Extensive experiments further demonstrate strong cross-modality transfer, robustness to linguistic variations and clinical language, as well as accurate instance-specific segmentation from real-world text. Code is available at: https://www.github.com/MIC-DKFZ/VoxTell
MeisenMeister: A Simple Two Stage Pipeline for Breast Cancer Classification on MRI
Oct 31, 2025Abstract:The ODELIA Breast MRI Challenge 2025 addresses a critical issue in breast cancer screening: improving early detection through more efficient and accurate interpretation of breast MRI scans. Even though methods for general-purpose whole-body lesion segmentation as well as multi-time-point analysis exist, breast cancer detection remains highly challenging, largely due to the limited availability of high-quality segmentation labels. Therefore, developing robust classification-based approaches is crucial for the future of early breast cancer detection, particularly in applications such as large-scale screening. In this write-up, we provide a comprehensive overview of our approach to the challenge. We begin by detailing the underlying concept and foundational assumptions that guided our work. We then describe the iterative development process, highlighting the key stages of experimentation, evaluation, and refinement that shaped the evolution of our solution. Finally, we present the reasoning and evidence that informed the design choices behind our final submission, with a focus on performance, robustness, and clinical relevance. We release our full implementation publicly at https://github.com/MIC-DKFZ/MeisenMeister
Temporal Flow Matching for Learning Spatio-Temporal Trajectories in 4D Longitudinal Medical Imaging
Aug 29, 2025



Abstract:Understanding temporal dynamics in medical imaging is crucial for applications such as disease progression modeling, treatment planning and anatomical development tracking. However, most deep learning methods either consider only single temporal contexts, or focus on tasks like classification or regression, limiting their ability for fine-grained spatial predictions. While some approaches have been explored, they are often limited to single timepoints, specific diseases or have other technical restrictions. To address this fundamental gap, we introduce Temporal Flow Matching (TFM), a unified generative trajectory method that (i) aims to learn the underlying temporal distribution, (ii) by design can fall back to a nearest image predictor, i.e. predicting the last context image (LCI), as a special case, and (iii) supports $3D$ volumes, multiple prior scans, and irregular sampling. Extensive benchmarks on three public longitudinal datasets show that TFM consistently surpasses spatio-temporal methods from natural imaging, establishing a new state-of-the-art and robust baseline for $4D$ medical image prediction.
Inclusive, Differentially Private Federated Learning for Clinical Data
May 28, 2025Abstract:Federated Learning (FL) offers a promising approach for training clinical AI models without centralizing sensitive patient data. However, its real-world adoption is hindered by challenges related to privacy, resource constraints, and compliance. Existing Differential Privacy (DP) approaches often apply uniform noise, which disproportionately degrades model performance, even among well-compliant institutions. In this work, we propose a novel compliance-aware FL framework that enhances DP by adaptively adjusting noise based on quantifiable client compliance scores. Additionally, we introduce a compliance scoring tool based on key healthcare and security standards to promote secure, inclusive, and equitable participation across diverse clinical settings. Extensive experiments on public datasets demonstrate that integrating under-resourced, less compliant clinics with highly regulated institutions yields accuracy improvements of up to 15% over traditional FL. This work advances FL by balancing privacy, compliance, and performance, making it a viable solution for real-world clinical workflows in global healthcare.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025


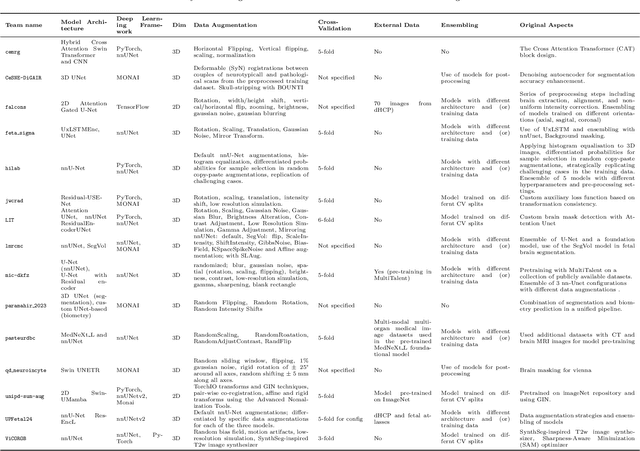
Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
nnLandmark: A Self-Configuring Method for 3D Medical Landmark Detection
Apr 10, 2025Abstract:Landmark detection plays a crucial role in medical imaging tasks that rely on precise spatial localization, including specific applications in diagnosis, treatment planning, image registration, and surgical navigation. However, manual annotation is labor-intensive and requires expert knowledge. While deep learning shows promise in automating this task, progress is hindered by limited public datasets, inconsistent benchmarks, and non-standardized baselines, restricting reproducibility, fair comparisons, and model generalizability. This work introduces nnLandmark, a self-configuring deep learning framework for 3D medical landmark detection, adapting nnU-Net to perform heatmap-based regression. By leveraging nnU-Net's automated configuration, nnLandmark eliminates the need for manual parameter tuning, offering out-of-the-box usability. It achieves state-of-the-art accuracy across two public datasets, with a mean radial error (MRE) of 1.5 mm on the Mandibular Molar Landmark (MML) dental CT dataset and 1.2 mm for anatomical fiducials on a brain MRI dataset (AFIDs), where nnLandmark aligns with the inter-rater variability of 1.5 mm. With its strong generalization, reproducibility, and ease of deployment, nnLandmark establishes a reliable baseline for 3D landmark detection, supporting research in anatomical localization and clinical workflows that depend on precise landmark identification. The code will be available soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge