Christian Wachinger
Cortex-Grounded Diffusion Models for Brain Image Generation
Jan 27, 2026Abstract:Synthetic neuroimaging data can mitigate critical limitations of real-world datasets, including the scarcity of rare phenotypes, domain shifts across scanners, and insufficient longitudinal coverage. However, existing generative models largely rely on weak conditioning signals, such as labels or text, which lack anatomical grounding and often produce biologically implausible outputs. To this end, we introduce Cor2Vox, a cortex-grounded generative framework for brain magnetic resonance image (MRI) synthesis that ties image generation to continuous structural priors of the cerebral cortex. It leverages high-resolution cortical surfaces to guide a 3D shape-to-image Brownian bridge diffusion process, enabling topologically faithful synthesis and precise control over underlying anatomies. To support the generation of new, realistic brain shapes, we developed a large-scale statistical shape model of cortical morphology derived from over 33,000 UK Biobank scans. We validated the fidelity of Cor2Vox based on traditional image quality metrics, advanced cortical surface reconstruction, and whole-brain segmentation quality, outperforming many baseline methods. Across three applications, namely (i) anatomically consistent synthesis, (ii) simulation of progressive gray matter atrophy, and (iii) harmonization of in-house frontotemporal dementia scans with public datasets, Cor2Vox preserved fine-grained cortical morphology at the sub-voxel level, exhibiting remarkable robustness to variations in cortical geometry and disease phenotype without retraining.
Template-Based Cortical Surface Reconstruction with Minimal Energy Deformation
Sep 18, 2025Abstract:Cortical surface reconstruction (CSR) from magnetic resonance imaging (MRI) is fundamental to neuroimage analysis, enabling morphological studies of the cerebral cortex and functional brain mapping. Recent advances in learning-based CSR have dramatically accelerated processing, allowing for reconstructions through the deformation of anatomical templates within seconds. However, ensuring the learned deformations are optimal in terms of deformation energy and consistent across training runs remains a particular challenge. In this work, we design a Minimal Energy Deformation (MED) loss, acting as a regularizer on the deformation trajectories and complementing the widely used Chamfer distance in CSR. We incorporate it into the recent V2C-Flow model and demonstrate considerable improvements in previously neglected training consistency and reproducibility without harming reconstruction accuracy and topological correctness.
Spherical Brownian Bridge Diffusion Models for Conditional Cortical Thickness Forecasting
Sep 10, 2025Abstract:Accurate forecasting of individualized, high-resolution cortical thickness (CTh) trajectories is essential for detecting subtle cortical changes, providing invaluable insights into neurodegenerative processes and facilitating earlier and more precise intervention strategies. However, CTh forecasting is a challenging task due to the intricate non-Euclidean geometry of the cerebral cortex and the need to integrate multi-modal data for subject-specific predictions. To address these challenges, we introduce the Spherical Brownian Bridge Diffusion Model (SBDM). Specifically, we propose a bidirectional conditional Brownian bridge diffusion process to forecast CTh trajectories at the vertex level of registered cortical surfaces. Our technical contribution includes a new denoising model, the conditional spherical U-Net (CoS-UNet), which combines spherical convolutions and dense cross-attention to integrate cortical surfaces and tabular conditions seamlessly. Compared to previous approaches, SBDM achieves significantly reduced prediction errors, as demonstrated by our experiments based on longitudinal datasets from the ADNI and OASIS. Additionally, we demonstrate SBDM's ability to generate individual factual and counterfactual CTh trajectories, offering a novel framework for exploring hypothetical scenarios of cortical development.
DISCO: Mitigating Bias in Deep Learning with Conditional Distance Correlation
Jun 13, 2025Abstract:During prediction tasks, models can use any signal they receive to come up with the final answer - including signals that are causally irrelevant. When predicting objects from images, for example, the lighting conditions could be correlated to different targets through selection bias, and an oblivious model might use these signals as shortcuts to discern between various objects. A predictor that uses lighting conditions instead of real object-specific details is obviously undesirable. To address this challenge, we introduce a standard anti-causal prediction model (SAM) that creates a causal framework for analyzing the information pathways influencing our predictor in anti-causal settings. We demonstrate that a classifier satisfying a specific conditional independence criterion will focus solely on the direct causal path from label to image, being counterfactually invariant to the remaining variables. Finally, we propose DISCO, a novel regularization strategy that uses conditional distance correlation to optimize for conditional independence in regression tasks. We can show that DISCO achieves competitive results in different bias mitigation experiments, deeming it a valid alternative to classical kernel-based methods.
MedBridge: Bridging Foundation Vision-Language Models to Medical Image Diagnosis
May 27, 2025Abstract:Recent vision-language foundation models deliver state-of-the-art results on natural image classification but falter on medical images due to pronounced domain shifts. At the same time, training a medical foundation model requires substantial resources, including extensive annotated data and high computational capacity. To bridge this gap with minimal overhead, we introduce MedBridge, a lightweight multimodal adaptation framework that re-purposes pretrained VLMs for accurate medical image diagnosis. MedBridge comprises three key components. First, a Focal Sampling module that extracts high-resolution local regions to capture subtle pathological features and compensate for the limited input resolution of general-purpose VLMs. Second, a Query Encoder (QEncoder) injects a small set of learnable queries that attend to the frozen feature maps of VLM, aligning them with medical semantics without retraining the entire backbone. Third, a Mixture of Experts mechanism, driven by learnable queries, harnesses the complementary strength of diverse VLMs to maximize diagnostic performance. We evaluate MedBridge on five medical imaging benchmarks across three key adaptation tasks, demonstrating its superior performance in both cross-domain and in-domain adaptation settings, even under varying levels of training data availability. Notably, MedBridge achieved over 6-15% improvement in AUC compared to state-of-the-art VLM adaptation methods in multi-label thoracic disease diagnosis, underscoring its effectiveness in leveraging foundation models for accurate and data-efficient medical diagnosis. Our code is available at https://github.com/ai-med/MedBridge.
3D Shape-to-Image Brownian Bridge Diffusion for Brain MRI Synthesis from Cortical Surfaces
Feb 18, 2025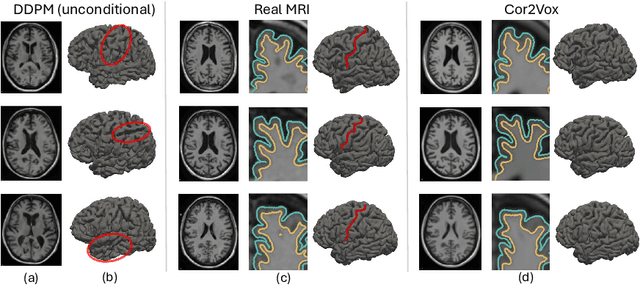
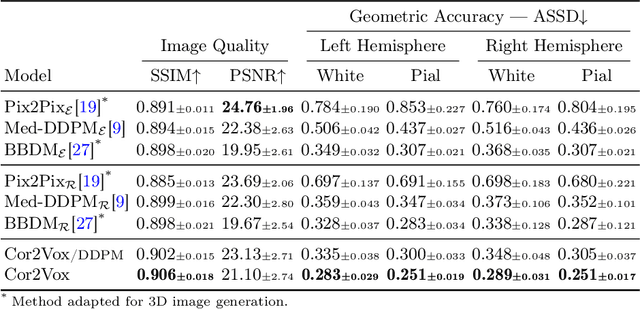
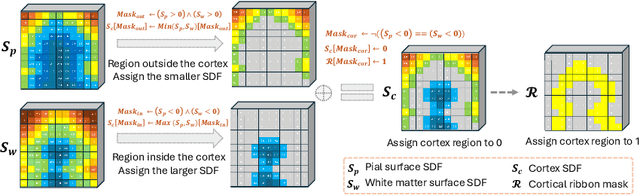
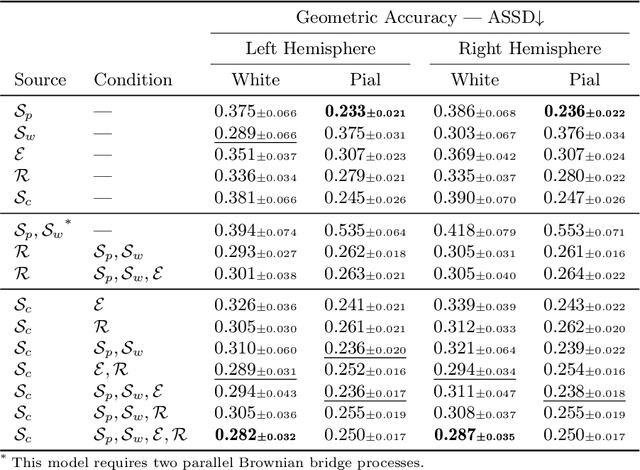
Abstract:Despite recent advances in medical image generation, existing methods struggle to produce anatomically plausible 3D structures. In synthetic brain magnetic resonance images (MRIs), characteristic fissures are often missing, and reconstructed cortical surfaces appear scattered rather than densely convoluted. To address this issue, we introduce Cor2Vox, the first diffusion model-based method that translates continuous cortical shape priors to synthetic brain MRIs. To achieve this, we leverage a Brownian bridge process which allows for direct structured mapping between shape contours and medical images. Specifically, we adapt the concept of the Brownian bridge diffusion model to 3D and extend it to embrace various complementary shape representations. Our experiments demonstrate significant improvements in the geometric accuracy of reconstructed structures compared to previous voxel-based approaches. Moreover, Cor2Vox excels in image quality and diversity, yielding high variation in non-target structures like the skull. Finally, we highlight the capability of our approach to simulate cortical atrophy at the sub-voxel level. Our code is available at https://github.com/ai-med/Cor2Vox.
WASUP: Interpretable Classification with Weight-Input Alignment and Class-Discriminative SUPports Vectors
Jan 28, 2025Abstract:The deployment of deep learning models in critical domains necessitates a balance between high accuracy and interpretability. We introduce WASUP, an inherently interpretable neural network that provides local and global explanations of its decision-making process. We prove that these explanations are faithful by fulfilling established axioms for explanations. Leveraging the concept of case-based reasoning, WASUP extracts class-representative support vectors from training images, ensuring they capture relevant features while suppressing irrelevant ones. Classification decisions are made by calculating and aggregating similarity scores between these support vectors and the input's latent feature vector. We employ B-Cos transformations, which align model weights with inputs to enable faithful mappings of latent features back to the input space, facilitating local explanations in addition to global explanations of case-based reasoning. We evaluate WASUP on three tasks: fine-grained classification on Stanford Dogs, multi-label classification on Pascal VOC, and pathology detection on the RSNA dataset. Results indicate that WASUP not only achieves competitive accuracy compared to state-of-the-art black-box models but also offers insightful explanations verified through theoretical analysis. Our findings underscore WASUP's potential for applications where understanding model decisions is as critical as the decisions themselves.
TimeFlow: Longitudinal Brain Image Registration and Aging Progression Analysis
Jan 15, 2025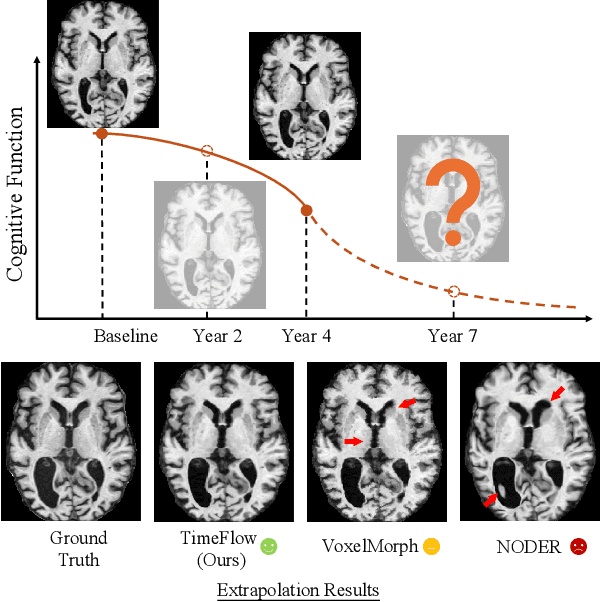


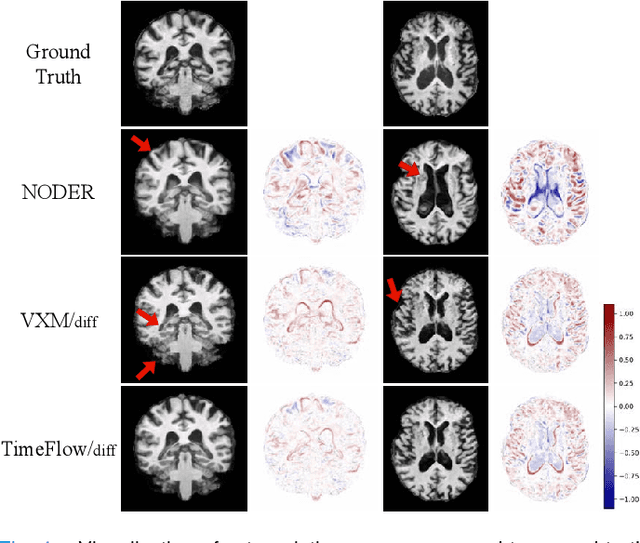
Abstract:Predicting future brain states is crucial for understanding healthy aging and neurodegenerative diseases. Longitudinal brain MRI registration, a cornerstone for such analyses, has long been limited by its inability to forecast future developments, reliance on extensive, dense longitudinal data, and the need to balance registration accuracy with temporal smoothness. In this work, we present \emph{TimeFlow}, a novel framework for longitudinal brain MRI registration that overcomes all these challenges. Leveraging a U-Net architecture with temporal conditioning inspired by diffusion models, TimeFlow enables accurate longitudinal registration and facilitates prospective analyses through future image prediction. Unlike traditional methods that depend on explicit smoothness regularizers and dense sequential data, TimeFlow achieves temporal consistency and continuity without these constraints. Experimental results highlight its superior performance in both future timepoint prediction and registration accuracy compared to state-of-the-art methods. Additionally, TimeFlow supports novel biological brain aging analyses, effectively differentiating neurodegenerative conditions from healthy aging. It eliminates the need for segmentation, thereby avoiding the challenges of non-trivial annotation and inconsistent segmentation errors. TimeFlow paves the way for accurate, data-efficient, and annotation-free prospective analyses of brain aging and chronic diseases.
MLV$^2$-Net: Rater-Based Majority-Label Voting for Consistent Meningeal Lymphatic Vessel Segmentation
Nov 13, 2024



Abstract:Meningeal lymphatic vessels (MLVs) are responsible for the drainage of waste products from the human brain. An impairment in their functionality has been associated with aging as well as brain disorders like multiple sclerosis and Alzheimer's disease. However, MLVs have only recently been described for the first time in magnetic resonance imaging (MRI), and their ramified structure renders manual segmentation particularly difficult. Further, as there is no consistent notion of their appearance, human-annotated MLV structures contain a high inter-rater variability that most automatic segmentation methods cannot take into account. In this work, we propose a new rater-aware training scheme for the popular nnU-Net model, and we explore rater-based ensembling strategies for accurate and consistent segmentation of MLVs. This enables us to boost nnU-Net's performance while obtaining explicit predictions in different annotation styles and a rater-based uncertainty estimation. Our final model, MLV$^2$-Net, achieves a Dice similarity coefficient of 0.806 with respect to the human reference standard. The model further matches the human inter-rater reliability and replicates age-related associations with MLV volume.
DiaMond: Dementia Diagnosis with Multi-Modal Vision Transformers Using MRI and PET
Oct 30, 2024Abstract:Diagnosing dementia, particularly for Alzheimer's Disease (AD) and frontotemporal dementia (FTD), is complex due to overlapping symptoms. While magnetic resonance imaging (MRI) and positron emission tomography (PET) data are critical for the diagnosis, integrating these modalities in deep learning faces challenges, often resulting in suboptimal performance compared to using single modalities. Moreover, the potential of multi-modal approaches in differential diagnosis, which holds significant clinical importance, remains largely unexplored. We propose a novel framework, DiaMond, to address these issues with vision Transformers to effectively integrate MRI and PET. DiaMond is equipped with self-attention and a novel bi-attention mechanism that synergistically combine MRI and PET, alongside a multi-modal normalization to reduce redundant dependency, thereby boosting the performance. DiaMond significantly outperforms existing multi-modal methods across various datasets, achieving a balanced accuracy of 92.4% in AD diagnosis, 65.2% for AD-MCI-CN classification, and 76.5% in differential diagnosis of AD and FTD. We also validated the robustness of DiaMond in a comprehensive ablation study. The code is available at https://github.com/ai-med/DiaMond.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge