Yu Li
Victor
ArcGen: Generalizing Neural Backdoor Detection Across Diverse Architectures
Dec 17, 2025Abstract:Backdoor attacks pose a significant threat to the security and reliability of deep learning models. To mitigate such attacks, one promising approach is to learn to extract features from the target model and use these features for backdoor detection. However, we discover that existing learning-based neural backdoor detection methods do not generalize well to new architectures not seen during the learning phase. In this paper, we analyze the root cause of this issue and propose a novel black-box neural backdoor detection method called ArcGen. Our method aims to obtain architecture-invariant model features, i.e., aligned features, for effective backdoor detection. Specifically, in contrast to existing methods directly using model outputs as model features, we introduce an additional alignment layer in the feature extraction function to further process these features. This reduces the direct influence of architecture information on the features. Then, we design two alignment losses to train the feature extraction function. These losses explicitly require that features from models with similar backdoor behaviors but different architectures are aligned at both the distribution and sample levels. With these techniques, our method demonstrates up to 42.5% improvements in detection performance (e.g., AUC) on unseen model architectures. This is based on a large-scale evaluation involving 16,896 models trained on diverse datasets, subjected to various backdoor attacks, and utilizing different model architectures. Our code is available at https://github.com/SeRAlab/ArcGen.
* 16 pages, 8 figures. This article was accepted by IEEE Transactions on Information Forensics and Security. DOI: 10.1109/TIFS.2025.3610254
OpenDataArena: A Fair and Open Arena for Benchmarking Post-Training Dataset Value
Dec 16, 2025



Abstract:The rapid evolution of Large Language Models (LLMs) is predicated on the quality and diversity of post-training datasets. However, a critical dichotomy persists: while models are rigorously benchmarked, the data fueling them remains a black box--characterized by opaque composition, uncertain provenance, and a lack of systematic evaluation. This opacity hinders reproducibility and obscures the causal link between data characteristics and model behaviors. To bridge this gap, we introduce OpenDataArena (ODA), a holistic and open platform designed to benchmark the intrinsic value of post-training data. ODA establishes a comprehensive ecosystem comprising four key pillars: (i) a unified training-evaluation pipeline that ensures fair, open comparisons across diverse models (e.g., Llama, Qwen) and domains; (ii) a multi-dimensional scoring framework that profiles data quality along tens of distinct axes; (iii) an interactive data lineage explorer to visualize dataset genealogy and dissect component sources; and (iv) a fully open-source toolkit for training, evaluation, and scoring to foster data research. Extensive experiments on ODA--covering over 120 training datasets across multiple domains on 22 benchmarks, validated by more than 600 training runs and 40 million processed data points--reveal non-trivial insights. Our analysis uncovers the inherent trade-offs between data complexity and task performance, identifies redundancy in popular benchmarks through lineage tracing, and maps the genealogical relationships across datasets. We release all results, tools, and configurations to democratize access to high-quality data evaluation. Rather than merely expanding a leaderboard, ODA envisions a shift from trial-and-error data curation to a principled science of Data-Centric AI, paving the way for rigorous studies on data mixing laws and the strategic composition of foundation models.
Investigating Data Pruning for Pretraining Biological Foundation Models at Scale
Dec 15, 2025Abstract:Biological foundation models (BioFMs), pretrained on large-scale biological sequences, have recently shown strong potential in providing meaningful representations for diverse downstream bioinformatics tasks. However, such models often rely on millions to billions of training sequences and billions of parameters, resulting in prohibitive computational costs and significant barriers to reproducibility and accessibility, particularly for academic labs. To address these challenges, we investigate the feasibility of data pruning for BioFM pretraining and propose a post-hoc influence-guided data pruning framework tailored to biological domains. Our approach introduces a subset-based self-influence formulation that enables efficient estimation of sample importance at low computational cost, and builds upon it two simple yet effective selection strategies, namely Top-k Influence (Top I) and Coverage-Centric Influence (CCI). We empirically validate our method on two representative BioFMs, RNA-FM and ESM-C. For RNA, our framework consistently outperforms random selection baselines under an extreme pruning rate of over 99 percent, demonstrating its effectiveness. Furthermore, we show the generalizability of our framework on protein-related tasks using ESM-C. In particular, our coreset even outperforms random subsets that are ten times larger in both RNA and protein settings, revealing substantial redundancy in biological sequence datasets. These findings underscore the potential of influence-guided data pruning to substantially reduce the computational cost of BioFM pretraining, paving the way for more efficient, accessible, and sustainable biological AI research.
Adaptive federated learning for ship detection across diverse satellite imagery sources
Dec 12, 2025Abstract:We investigate the application of Federated Learning (FL) for ship detection across diverse satellite datasets, offering a privacy-preserving solution that eliminates the need for data sharing or centralized collection. This approach is particularly advantageous for handling commercial satellite imagery or sensitive ship annotations. Four FL models including FedAvg, FedProx, FedOpt, and FedMedian, are evaluated and compared to a local training baseline, where the YOLOv8 ship detection model is independently trained on each dataset without sharing learned parameters. The results reveal that FL models substantially improve detection accuracy over training on smaller local datasets and achieve performance levels close to global training that uses all datasets during the training. Furthermore, the study underscores the importance of selecting appropriate FL configurations, such as the number of communication rounds and local training epochs, to optimize detection precision while maintaining computational efficiency.
C$^3$TG: Conflict-aware, Composite, and Collaborative Controlled Text Generation
Nov 16, 2025Abstract:Recent advancements in large language models (LLMs) have demonstrated remarkable text generation capabilities. However, controlling specific attributes of generated text remains challenging without architectural modifications or extensive fine-tuning. Current methods typically toggle a single, basic attribute but struggle with precise multi-attribute control. In scenarios where attribute requirements conflict, existing methods lack coordination mechanisms, causing interference between desired attributes. Furthermore, these methods fail to incorporate iterative optimization processes in the controlled generation pipeline. To address these limitations, we propose Conflict-aware, Composite, and Collaborative Controlled Text Generation (C$^3$TG), a two-phase framework for fine-grained, multi-dimensional text attribute control. During generation, C$^3$TG selectively pairs the LLM with the required attribute classifiers from the 17 available dimensions and employs weighted KL-divergence to adjust token probabilities. The optimization phase then leverages an energy function combining classifier scores and penalty terms to resolve attribute conflicts through iterative feedback, enabling precise control over multiple dimensions simultaneously while preserving natural text flow. Experiments show that C$^3$TG significantly outperforms baselines across multiple metrics including attribute accuracy, linguistic fluency, and output diversity, while simultaneously reducing toxicity. These results establish C$^3$TG as an effective and flexible solution for multi-dimensional text attribute control that requires no costly model modifications.
From Static Structures to Ensembles: Studying and Harnessing Protein Structure Tokenization
Nov 13, 2025Abstract:Protein structure tokenization converts 3D structures into discrete or vectorized representations, enabling the integration of structural and sequence data. Despite many recent works on structure tokenization, the properties of the underlying discrete representations are not well understood. In this work, we first demonstrate that the successful utilization of structural tokens in a language model for structure prediction depends on using rich, pre-trained sequence embeddings to bridge the semantic gap between the sequence and structural "language". The analysis of the structural vocabulary itself then reveals significant semantic redundancy, where multiple distinct tokens correspond to nearly identical local geometries, acting as "structural synonyms". This redundancy, rather than being a flaw, can be exploited with a simple "synonym swap" strategy to generate diverse conformational ensembles by perturbing a predicted structure with its structural synonyms. This computationally lightweight method accurately recapitulates protein flexibility, performing competitively with state-of-the-art models. Our study provides fundamental insights into the nature of discrete protein structure representations and introduces a powerful, near-instantaneous method for modeling protein dynamics. Source code is available in https://github.com/IDEA-XL/TokenMD.
AgentExpt: Automating AI Experiment Design with LLM-based Resource Retrieval Agent
Nov 07, 2025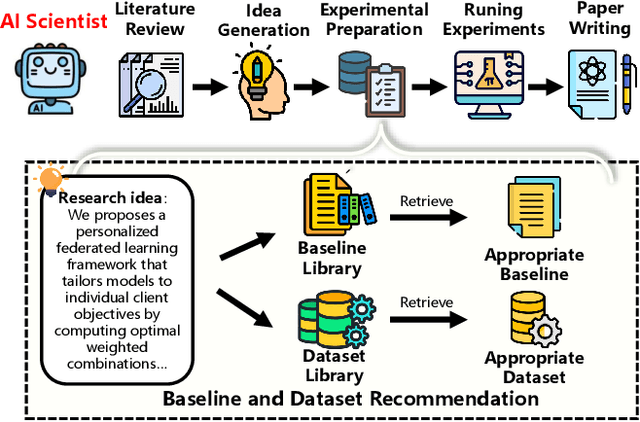
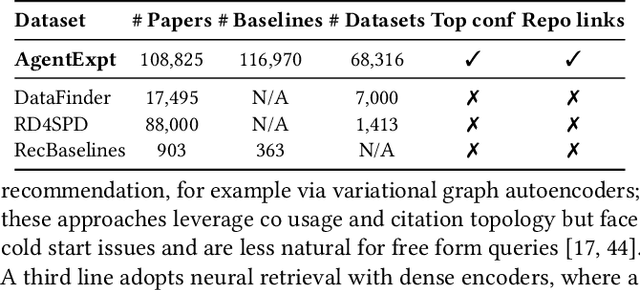
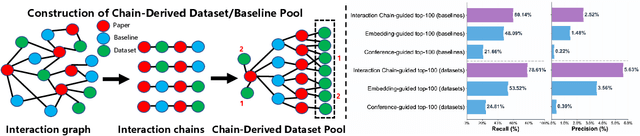
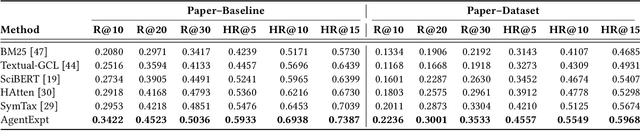
Abstract:Large language model agents are becoming increasingly capable at web-centric tasks such as information retrieval, complex reasoning. These emerging capabilities have given rise to surge research interests in developing LLM agent for facilitating scientific quest. One key application in AI research is to automate experiment design through agentic dataset and baseline retrieval. However, prior efforts suffer from limited data coverage, as recommendation datasets primarily harvest candidates from public portals and omit many datasets actually used in published papers, and from an overreliance on content similarity that biases model toward superficial similarity and overlooks experimental suitability. Harnessing collective perception embedded in the baseline and dataset citation network, we present a comprehensive framework for baseline and dataset recommendation. First, we design an automated data-collection pipeline that links roughly one hundred thousand accepted papers to the baselines and datasets they actually used. Second, we propose a collective perception enhanced retriever. To represent the position of each dataset or baseline within the scholarly network, it concatenates self-descriptions with aggregated citation contexts. To achieve efficient candidate recall, we finetune an embedding model on these representations. Finally, we develop a reasoning-augmented reranker that exact interaction chains to construct explicit reasoning chains and finetunes a large language model to produce interpretable justifications and refined rankings. The dataset we curated covers 85\% of the datasets and baselines used at top AI conferences over the past five years. On our dataset, the proposed method outperforms the strongest prior baseline with average gains of +5.85\% in Recall@20, +8.30\% in HitRate@5. Taken together, our results advance reliable, interpretable automation of experimental design.
Inverse Knowledge Search over Verifiable Reasoning: Synthesizing a Scientific Encyclopedia from a Long Chains-of-Thought Knowledge Base
Oct 30, 2025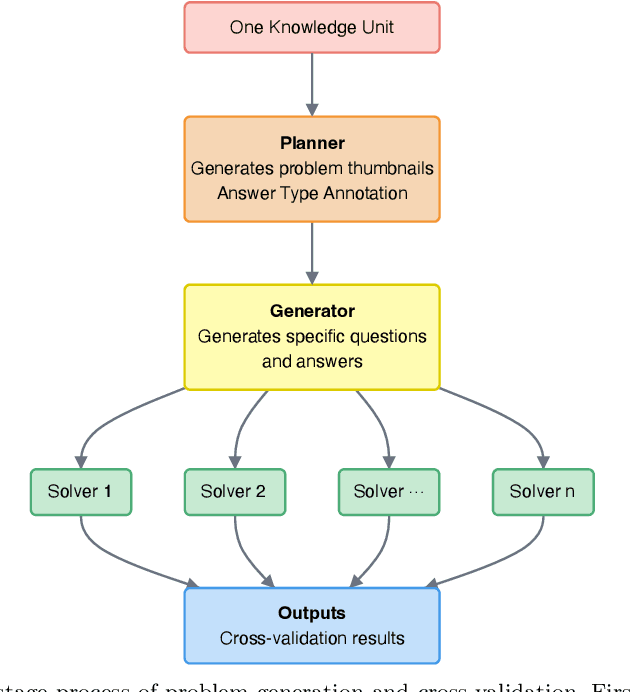
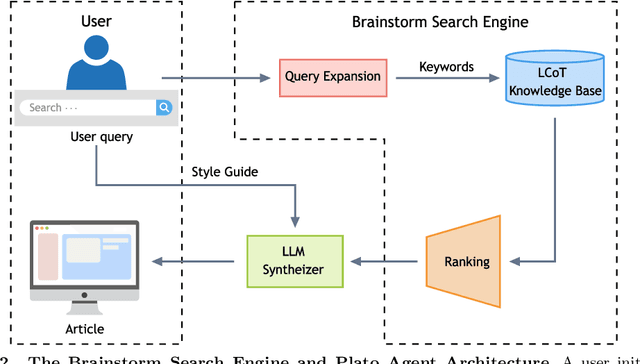
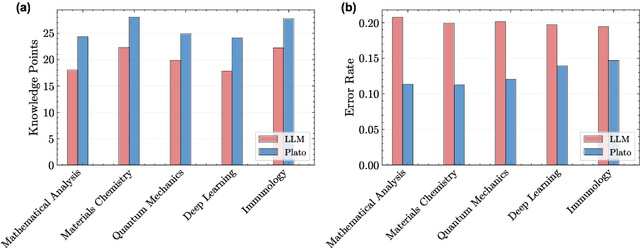
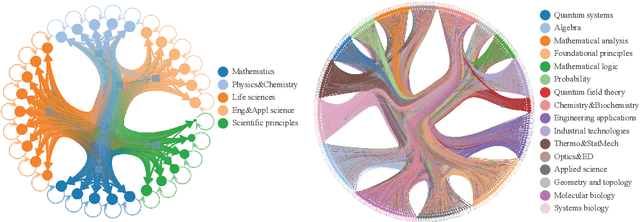
Abstract:Most scientific materials compress reasoning, presenting conclusions while omitting the derivational chains that justify them. This compression hinders verification by lacking explicit, step-wise justifications and inhibits cross-domain links by collapsing the very pathways that establish the logical and causal connections between concepts. We introduce a scalable framework that decompresses scientific reasoning, constructing a verifiable Long Chain-of-Thought (LCoT) knowledge base and projecting it into an emergent encyclopedia, SciencePedia. Our pipeline operationalizes an endpoint-driven, reductionist strategy: a Socratic agent, guided by a curriculum of around 200 courses, generates approximately 3 million first-principles questions. To ensure high fidelity, multiple independent solver models generate LCoTs, which are then rigorously filtered by prompt sanitization and cross-model answer consensus, retaining only those with verifiable endpoints. This verified corpus powers the Brainstorm Search Engine, which performs inverse knowledge search -- retrieving diverse, first-principles derivations that culminate in a target concept. This engine, in turn, feeds the Plato synthesizer, which narrates these verified chains into coherent articles. The initial SciencePedia comprises approximately 200,000 fine-grained entries spanning mathematics, physics, chemistry, biology, engineering, and computation. In evaluations across six disciplines, Plato-synthesized articles (conditioned on retrieved LCoTs) exhibit substantially higher knowledge-point density and significantly lower factual error rates than an equally-prompted baseline without retrieval (as judged by an external LLM). Built on this verifiable LCoT knowledge base, this reasoning-centric approach enables trustworthy, cross-domain scientific synthesis at scale and establishes the foundation for an ever-expanding encyclopedia.
MelCap: A Unified Single-Codebook Neural Codec for High-Fidelity Audio Compression
Oct 02, 2025Abstract:Neural audio codecs have recently emerged as powerful tools for high-quality and low-bitrate audio compression, leveraging deep generative models to learn latent representations of audio signals. However, existing approaches either rely on a single quantizer that only processes speech domain, or on multiple quantizers that are not well suited for downstream tasks. To address this issue, we propose MelCap, a unified "one-codebook-for-all" neural codec that effectively handles speech, music, and general sound. By decomposing audio reconstruction into two stages, our method preserves more acoustic details than previous single-codebook approaches, while achieving performance comparable to mainstream multi-codebook methods. In the first stage, audio is transformed into mel-spectrograms, which are compressed and quantized into compact single tokens using a 2D tokenizer. A perceptual loss is further applied to mitigate the over-smoothing artifacts observed in spectrogram reconstruction. In the second stage, a Vocoder recovers waveforms from the mel discrete tokens in a single forward pass, enabling real-time decoding. Both objective and subjective evaluations demonstrate that MelCap achieves quality on comparable to state-of-the-art multi-codebook codecs, while retaining the computational simplicity of a single-codebook design, thereby providing an effective representation for downstream tasks.
Prompt2Auto: From Motion Prompt to Automated Control via Geometry-Invariant One-Shot Gaussian Process Learning
Sep 17, 2025Abstract:Learning from demonstration allows robots to acquire complex skills from human demonstrations, but conventional approaches often require large datasets and fail to generalize across coordinate transformations. In this paper, we propose Prompt2Auto, a geometry-invariant one-shot Gaussian process (GeoGP) learning framework that enables robots to perform human-guided automated control from a single motion prompt. A dataset-construction strategy based on coordinate transformations is introduced that enforces invariance to translation, rotation, and scaling, while supporting multi-step predictions. Moreover, GeoGP is robust to variations in the user's motion prompt and supports multi-skill autonomy. We validate the proposed approach through numerical simulations with the designed user graphical interface and two real-world robotic experiments, which demonstrate that the proposed method is effective, generalizes across tasks, and significantly reduces the demonstration burden. Project page is available at: https://prompt2auto.github.io
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge