Mao Su
An Agentic Framework for Autonomous Materials Computation
Dec 22, 2025



Abstract:Large Language Models (LLMs) have emerged as powerful tools for accelerating scientific discovery, yet their static knowledge and hallucination issues hinder autonomous research applications. Recent advances integrate LLMs into agentic frameworks, enabling retrieval, reasoning, and tool use for complex scientific workflows. Here, we present a domain-specialized agent designed for reliable automation of first-principles materials computations. By embedding domain expertise, the agent ensures physically coherent multi-step workflows and consistently selects convergent, well-posed parameters, thereby enabling reliable end-to-end computational execution. A new benchmark of diverse computational tasks demonstrates that our system significantly outperforms standalone LLMs in both accuracy and robustness. This work establishes a verifiable foundation for autonomous computational experimentation and represents a key step toward fully automated scientific discovery.
Probing Scientific General Intelligence of LLMs with Scientist-Aligned Workflows
Dec 18, 2025Abstract:Despite advances in scientific AI, a coherent framework for Scientific General Intelligence (SGI)-the ability to autonomously conceive, investigate, and reason across scientific domains-remains lacking. We present an operational SGI definition grounded in the Practical Inquiry Model (PIM: Deliberation, Conception, Action, Perception) and operationalize it via four scientist-aligned tasks: deep research, idea generation, dry/wet experiments, and experimental reasoning. SGI-Bench comprises over 1,000 expert-curated, cross-disciplinary samples inspired by Science's 125 Big Questions, enabling systematic evaluation of state-of-the-art LLMs. Results reveal gaps: low exact match (10--20%) in deep research despite step-level alignment; ideas lacking feasibility and detail; high code executability but low execution result accuracy in dry experiments; low sequence fidelity in wet protocols; and persistent multimodal comparative-reasoning challenges. We further introduce Test-Time Reinforcement Learning (TTRL), which optimizes retrieval-augmented novelty rewards at inference, enhancing hypothesis novelty without reference answer. Together, our PIM-grounded definition, workflow-centric benchmark, and empirical insights establish a foundation for AI systems that genuinely participate in scientific discovery.
ATLAS: A High-Difficulty, Multidisciplinary Benchmark for Frontier Scientific Reasoning
Nov 18, 2025Abstract:The rapid advancement of Large Language Models (LLMs) has led to performance saturation on many established benchmarks, questioning their ability to distinguish frontier models. Concurrently, existing high-difficulty benchmarks often suffer from narrow disciplinary focus, oversimplified answer formats, and vulnerability to data contamination, creating a fidelity gap with real-world scientific inquiry. To address these challenges, we introduce ATLAS (AGI-Oriented Testbed for Logical Application in Science), a large-scale, high-difficulty, and cross-disciplinary evaluation suite composed of approximately 800 original problems. Developed by domain experts (PhD-level and above), ATLAS spans seven core scientific fields: mathematics, physics, chemistry, biology, computer science, earth science, and materials science. Its key features include: (1) High Originality and Contamination Resistance, with all questions newly created or substantially adapted to prevent test data leakage; (2) Cross-Disciplinary Focus, designed to assess models' ability to integrate knowledge and reason across scientific domains; (3) High-Fidelity Answers, prioritizing complex, open-ended answers involving multi-step reasoning and LaTeX-formatted expressions over simple multiple-choice questions; and (4) Rigorous Quality Control, employing a multi-stage process of expert peer review and adversarial testing to ensure question difficulty, scientific value, and correctness. We also propose a robust evaluation paradigm using a panel of LLM judges for automated, nuanced assessment of complex answers. Preliminary results on leading models demonstrate ATLAS's effectiveness in differentiating their advanced scientific reasoning capabilities. We plan to develop ATLAS into a long-term, open, community-driven platform to provide a reliable "ruler" for progress toward Artificial General Intelligence.
ChemBOMAS: Accelerated BO in Chemistry with LLM-Enhanced Multi-Agent System
Sep 10, 2025Abstract:The efficiency of Bayesian optimization (BO) in chemistry is often hindered by sparse experimental data and complex reaction mechanisms. To overcome these limitations, we introduce ChemBOMAS, a new framework named LLM-Enhanced Multi-Agent System for accelerating BO in chemistry. ChemBOMAS's optimization process is enhanced by LLMs and synergistically employs two strategies: knowledge-driven coarse-grained optimization and data-driven fine-grained optimization. First, in the knowledge-driven coarse-grained optimization stage, LLMs intelligently decompose the vast search space by reasoning over existing chemical knowledge to identify promising candidate regions. Subsequently, in the data-driven fine-grained optimization stage, LLMs enhance the BO process within these candidate regions by generating pseudo-data points, thereby improving data utilization efficiency and accelerating convergence. Benchmark evaluations** further confirm that ChemBOMAS significantly enhances optimization effectiveness and efficiency compared to various BO algorithms. Importantly, the practical utility of ChemBOMAS was validated through wet-lab experiments conducted under pharmaceutical industry protocols, targeting conditional optimization for a previously unreported and challenging chemical reaction. In the wet experiment, ChemBOMAS achieved an optimal objective value of 96%. This was substantially higher than the 15% achieved by domain experts. This real-world success, together with strong performance on benchmark evaluations, highlights ChemBOMAS as a powerful tool to accelerate chemical discovery.
CMPhysBench: A Benchmark for Evaluating Large Language Models in Condensed Matter Physics
Aug 25, 2025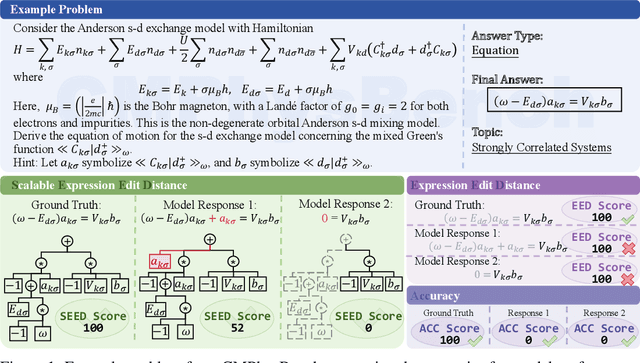
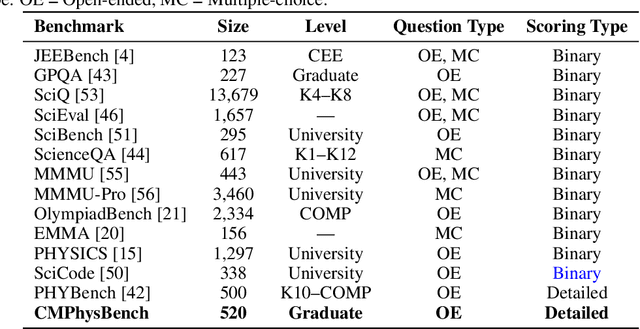
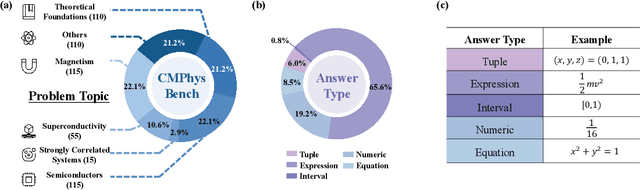
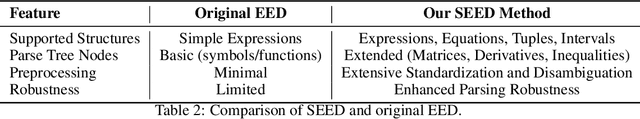
Abstract:We introduce CMPhysBench, designed to assess the proficiency of Large Language Models (LLMs) in Condensed Matter Physics, as a novel Benchmark. CMPhysBench is composed of more than 520 graduate-level meticulously curated questions covering both representative subfields and foundational theoretical frameworks of condensed matter physics, such as magnetism, superconductivity, strongly correlated systems, etc. To ensure a deep understanding of the problem-solving process,we focus exclusively on calculation problems, requiring LLMs to independently generate comprehensive solutions. Meanwhile, leveraging tree-based representations of expressions, we introduce the Scalable Expression Edit Distance (SEED) score, which provides fine-grained (non-binary) partial credit and yields a more accurate assessment of similarity between prediction and ground-truth. Our results show that even the best models, Grok-4, reach only 36 average SEED score and 28% accuracy on CMPhysBench, underscoring a significant capability gap, especially for this practical and frontier domain relative to traditional physics. The code anddataset are publicly available at https://github.com/CMPhysBench/CMPhysBench.
Iterative Pretraining Framework for Interatomic Potentials
Jul 27, 2025Abstract:Machine learning interatomic potentials (MLIPs) enable efficient molecular dynamics (MD) simulations with ab initio accuracy and have been applied across various domains in physical science. However, their performance often relies on large-scale labeled training data. While existing pretraining strategies can improve model performance, they often suffer from a mismatch between the objectives of pretraining and downstream tasks or rely on extensive labeled datasets and increasingly complex architectures to achieve broad generalization. To address these challenges, we propose Iterative Pretraining for Interatomic Potentials (IPIP), a framework designed to iteratively improve the predictive performance of MLIP models. IPIP incorporates a forgetting mechanism to prevent iterative training from converging to suboptimal local minima. Unlike general-purpose foundation models, which frequently underperform on specialized tasks due to a trade-off between generality and system-specific accuracy, IPIP achieves higher accuracy and efficiency using lightweight architectures. Compared to general-purpose force fields, this approach achieves over 80% reduction in prediction error and up to 4x speedup in the challenging Mo-S-O system, enabling fast and accurate simulations.
ChemVLM: Exploring the Power of Multimodal Large Language Models in Chemistry Area
Aug 16, 2024



Abstract:Large Language Models (LLMs) have achieved remarkable success and have been applied across various scientific fields, including chemistry. However, many chemical tasks require the processing of visual information, which cannot be successfully handled by existing chemical LLMs. This brings a growing need for models capable of integrating multimodal information in the chemical domain. In this paper, we introduce \textbf{ChemVLM}, an open-source chemical multimodal large language model specifically designed for chemical applications. ChemVLM is trained on a carefully curated bilingual multimodal dataset that enhances its ability to understand both textual and visual chemical information, including molecular structures, reactions, and chemistry examination questions. We develop three datasets for comprehensive evaluation, tailored to Chemical Optical Character Recognition (OCR), Multimodal Chemical Reasoning (MMCR), and Multimodal Molecule Understanding tasks. We benchmark ChemVLM against a range of open-source and proprietary multimodal large language models on various tasks. Experimental results demonstrate that ChemVLM achieves competitive performance across all evaluated tasks. Our model can be found at https://huggingface.co/AI4Chem/ChemVLM-26B.
Seeing and Understanding: Bridging Vision with Chemical Knowledge Via ChemVLM
Aug 14, 2024



Abstract:In this technical report, we propose ChemVLM, the first open-source multimodal large language model dedicated to the fields of chemistry, designed to address the incompatibility between chemical image understanding and text analysis. Built upon the VIT-MLP-LLM architecture, we leverage ChemLLM-20B as the foundational large model, endowing our model with robust capabilities in understanding and utilizing chemical text knowledge. Additionally, we employ InternVIT-6B as a powerful image encoder. We have curated high-quality data from the chemical domain, including molecules, reaction formulas, and chemistry examination data, and compiled these into a bilingual multimodal question-answering dataset. We test the performance of our model on multiple open-source benchmarks and three custom evaluation sets. Experimental results demonstrate that our model achieves excellent performance, securing state-of-the-art results in five out of six involved tasks. Our model can be found at https://huggingface.co/AI4Chem/ChemVLM-26B.
Physical formula enhanced multi-task learning for pharmacokinetics prediction
Apr 16, 2024



Abstract:Artificial intelligence (AI) technology has demonstrated remarkable potential in drug dis-covery, where pharmacokinetics plays a crucial role in determining the dosage, safety, and efficacy of new drugs. A major challenge for AI-driven drug discovery (AIDD) is the scarcity of high-quality data, which often requires extensive wet-lab work. A typical example of this is pharmacokinetic experiments. In this work, we develop a physical formula enhanced mul-ti-task learning (PEMAL) method that predicts four key parameters of pharmacokinetics simultaneously. By incorporating physical formulas into the multi-task framework, PEMAL facilitates effective knowledge sharing and target alignment among the pharmacokinetic parameters, thereby enhancing the accuracy of prediction. Our experiments reveal that PEMAL significantly lowers the data demand, compared to typical Graph Neural Networks. Moreover, we demonstrate that PEMAL enhances the robustness to noise, an advantage that conventional Neural Networks do not possess. Another advantage of PEMAL is its high flexibility, which can be potentially applied to other multi-task machine learning scenarios. Overall, our work illustrates the benefits and potential of using PEMAL in AIDD and other scenarios with data scarcity and noise.
ChemLLM: A Chemical Large Language Model
Feb 10, 2024



Abstract:Large language models (LLMs) have made impressive progress in chemistry applications, including molecular property prediction, molecular generation, experimental protocol design, etc. However, the community lacks a dialogue-based model specifically designed for chemistry. The challenge arises from the fact that most chemical data and scientific knowledge are primarily stored in structured databases, and the direct use of these structured data compromises the model's ability to maintain coherent dialogue. To tackle this issue, we develop a novel template-based instruction construction method that transforms structured knowledge into plain dialogue, making it suitable for language model training. By leveraging this approach, we develop ChemLLM, the first large language model dedicated to chemistry, capable of performing various tasks across chemical disciplines with smooth dialogue interaction. ChemLLM beats GPT-3.5 on all three principal tasks in chemistry, i.e., name conversion, molecular caption, and reaction prediction, and surpasses GPT-4 on two of them. Remarkably, ChemLLM also shows exceptional adaptability to related mathematical and physical tasks despite being trained mainly on chemical-centric corpora. Furthermore, ChemLLM demonstrates proficiency in specialized NLP tasks within chemistry, such as literature translation and cheminformatic programming. ChemLLM opens up a new avenue for exploration within chemical studies, while our method of integrating structured chemical knowledge into dialogue systems sets a new frontier for developing LLMs across various scientific fields. Codes, Datasets, and Model weights are publicly accessible at hf.co/AI4Chem/ChemLLM-7B-Chat.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge