Xiangxin Zhou
Rethinking the Trust Region in LLM Reinforcement Learning
Feb 04, 2026Abstract:Reinforcement learning (RL) has become a cornerstone for fine-tuning Large Language Models (LLMs), with Proximal Policy Optimization (PPO) serving as the de facto standard algorithm. Despite its ubiquity, we argue that the core ratio clipping mechanism in PPO is structurally ill-suited for the large vocabularies inherent to LLMs. PPO constrains policy updates based on the probability ratio of sampled tokens, which serves as a noisy single-sample Monte Carlo estimate of the true policy divergence. This creates a sub-optimal learning dynamic: updates to low-probability tokens are aggressively over-penalized, while potentially catastrophic shifts in high-probability tokens are under-constrained, leading to training inefficiency and instability. To address this, we propose Divergence Proximal Policy Optimization (DPPO), which substitutes heuristic clipping with a more principled constraint based on a direct estimate of policy divergence (e.g., Total Variation or KL). To avoid huge memory footprint, we introduce the efficient Binary and Top-K approximations to capture the essential divergence with negligible overhead. Extensive empirical evaluations demonstrate that DPPO achieves superior training stability and efficiency compared to existing methods, offering a more robust foundation for RL-based LLM fine-tuning.
Defeating the Training-Inference Mismatch via FP16
Oct 30, 2025Abstract:Reinforcement learning (RL) fine-tuning of large language models (LLMs) often suffers from instability due to the numerical mismatch between the training and inference policies. While prior work has attempted to mitigate this issue through algorithmic corrections or engineering alignments, we show that its root cause lies in the floating point precision itself. The widely adopted BF16, despite its large dynamic range, introduces large rounding errors that breaks the consistency between training and inference. In this work, we demonstrate that simply reverting to \textbf{FP16} effectively eliminates this mismatch. The change is simple, fully supported by modern frameworks with only a few lines of code change, and requires no modification to the model architecture or learning algorithm. Our results suggest that using FP16 uniformly yields more stable optimization, faster convergence, and stronger performance across diverse tasks, algorithms and frameworks. We hope these findings motivate a broader reconsideration of precision trade-offs in RL fine-tuning.
Variational Reasoning for Language Models
Sep 26, 2025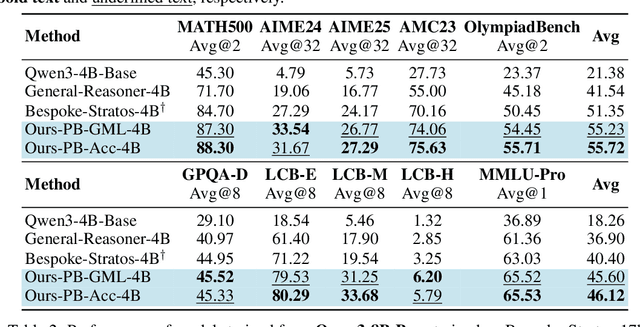
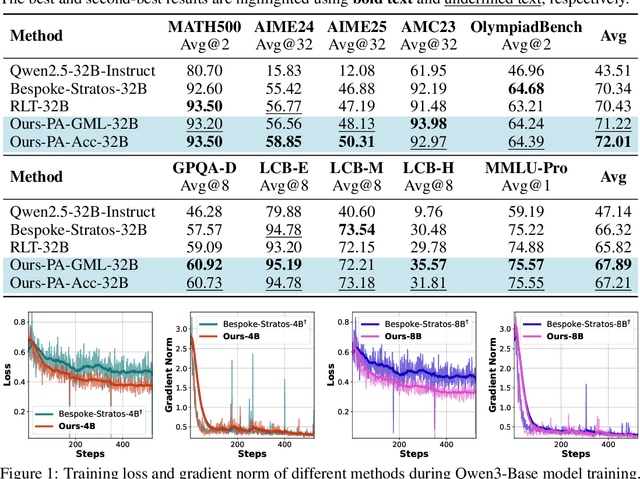
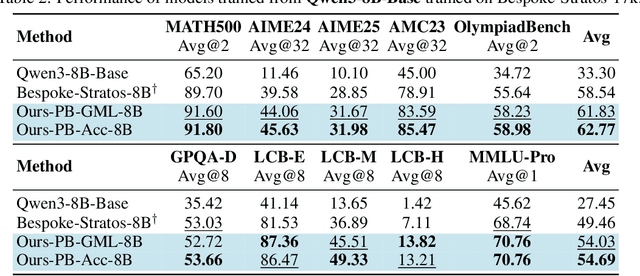
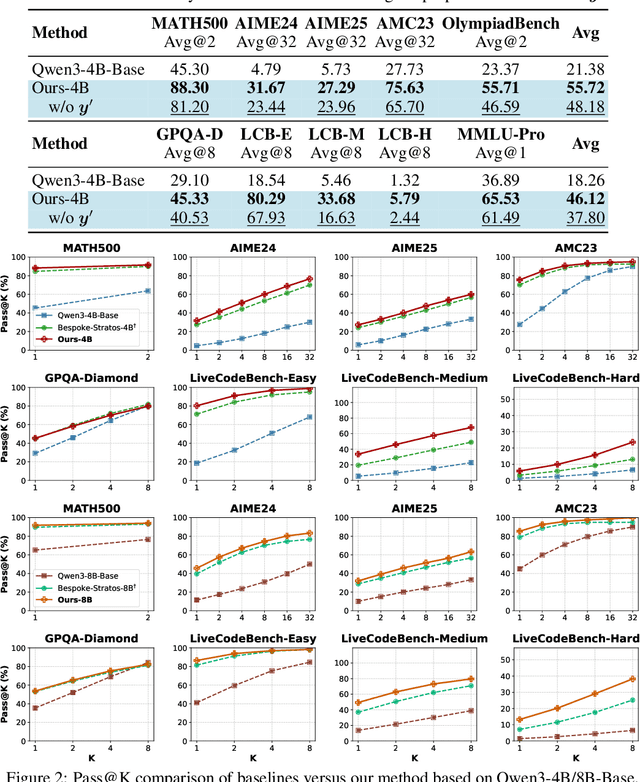
Abstract:We introduce a variational reasoning framework for language models that treats thinking traces as latent variables and optimizes them through variational inference. Starting from the evidence lower bound (ELBO), we extend it to a multi-trace objective for tighter bounds and propose a forward-KL formulation that stabilizes the training of the variational posterior. We further show that rejection sampling finetuning and binary-reward RL, including GRPO, can be interpreted as local forward-KL objectives, where an implicit weighting by model accuracy naturally arises from the derivation and reveals a previously unnoticed bias toward easier questions. We empirically validate our method on the Qwen 2.5 and Qwen 3 model families across a wide range of reasoning tasks. Overall, our work provides a principled probabilistic perspective that unifies variational inference with RL-style methods and yields stable objectives for improving the reasoning ability of language models. Our code is available at https://github.com/sail-sg/variational-reasoning.
ChemBOMAS: Accelerated BO in Chemistry with LLM-Enhanced Multi-Agent System
Sep 10, 2025Abstract:The efficiency of Bayesian optimization (BO) in chemistry is often hindered by sparse experimental data and complex reaction mechanisms. To overcome these limitations, we introduce ChemBOMAS, a new framework named LLM-Enhanced Multi-Agent System for accelerating BO in chemistry. ChemBOMAS's optimization process is enhanced by LLMs and synergistically employs two strategies: knowledge-driven coarse-grained optimization and data-driven fine-grained optimization. First, in the knowledge-driven coarse-grained optimization stage, LLMs intelligently decompose the vast search space by reasoning over existing chemical knowledge to identify promising candidate regions. Subsequently, in the data-driven fine-grained optimization stage, LLMs enhance the BO process within these candidate regions by generating pseudo-data points, thereby improving data utilization efficiency and accelerating convergence. Benchmark evaluations** further confirm that ChemBOMAS significantly enhances optimization effectiveness and efficiency compared to various BO algorithms. Importantly, the practical utility of ChemBOMAS was validated through wet-lab experiments conducted under pharmaceutical industry protocols, targeting conditional optimization for a previously unreported and challenging chemical reaction. In the wet experiment, ChemBOMAS achieved an optimal objective value of 96%. This was substantially higher than the 15% achieved by domain experts. This real-world success, together with strong performance on benchmark evaluations, highlights ChemBOMAS as a powerful tool to accelerate chemical discovery.
OS Agents: A Survey on MLLM-based Agents for General Computing Devices Use
Aug 06, 2025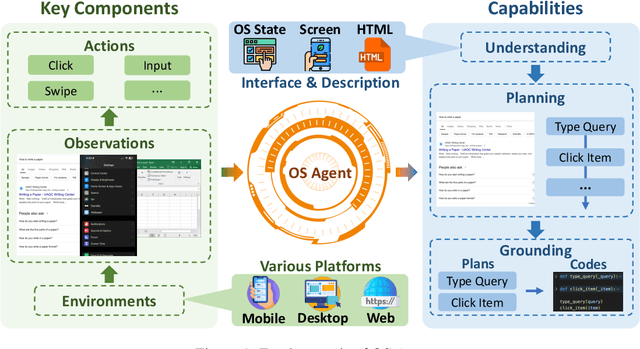
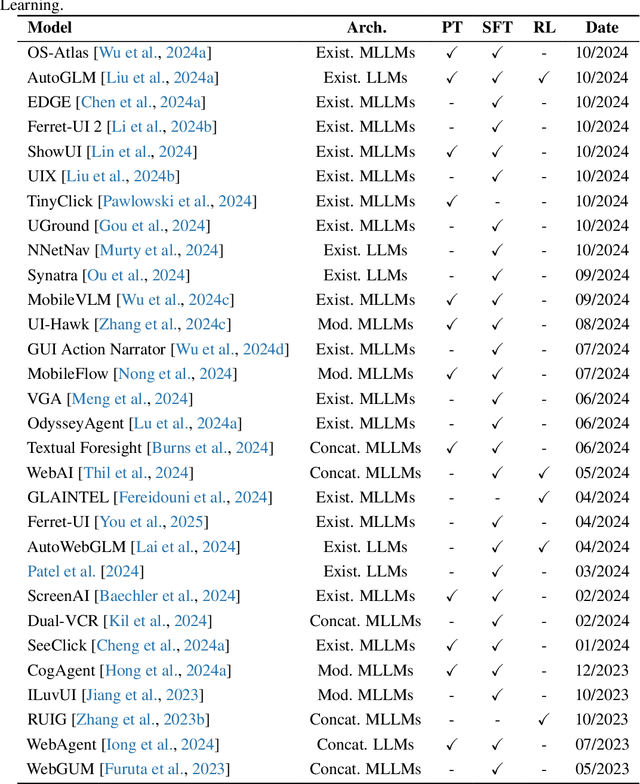
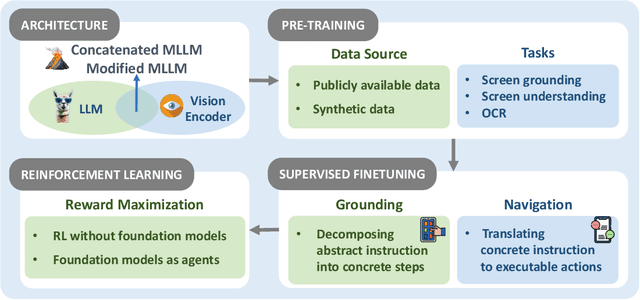
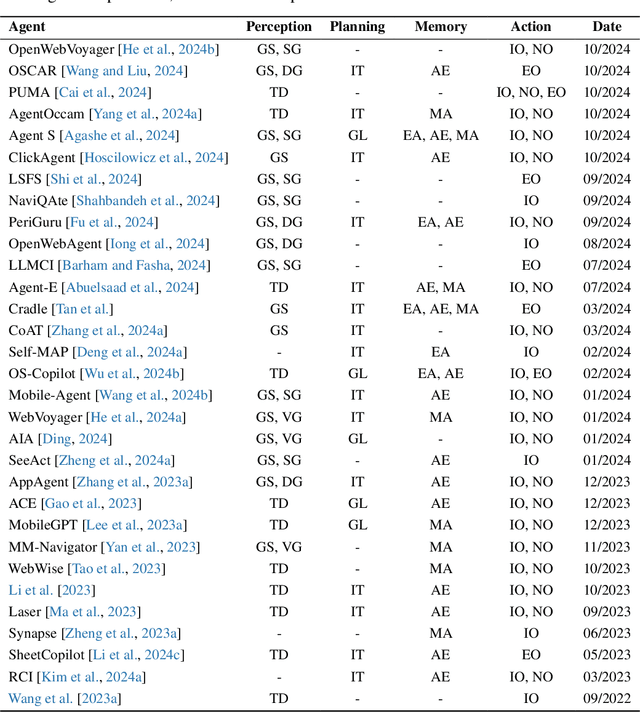
Abstract:The dream to create AI assistants as capable and versatile as the fictional J.A.R.V.I.S from Iron Man has long captivated imaginations. With the evolution of (multi-modal) large language models ((M)LLMs), this dream is closer to reality, as (M)LLM-based Agents using computing devices (e.g., computers and mobile phones) by operating within the environments and interfaces (e.g., Graphical User Interface (GUI)) provided by operating systems (OS) to automate tasks have significantly advanced. This paper presents a comprehensive survey of these advanced agents, designated as OS Agents. We begin by elucidating the fundamentals of OS Agents, exploring their key components including the environment, observation space, and action space, and outlining essential capabilities such as understanding, planning, and grounding. We then examine methodologies for constructing OS Agents, focusing on domain-specific foundation models and agent frameworks. A detailed review of evaluation protocols and benchmarks highlights how OS Agents are assessed across diverse tasks. Finally, we discuss current challenges and identify promising directions for future research, including safety and privacy, personalization and self-evolution. This survey aims to consolidate the state of OS Agents research, providing insights to guide both academic inquiry and industrial development. An open-source GitHub repository is maintained as a dynamic resource to foster further innovation in this field. We present a 9-page version of our work, accepted by ACL 2025, to provide a concise overview to the domain.
Reinforcing General Reasoning without Verifiers
May 27, 2025Abstract:The recent paradigm shift towards training large language models (LLMs) using DeepSeek-R1-Zero-style reinforcement learning (RL) on verifiable rewards has led to impressive advancements in code and mathematical reasoning. However, this methodology is limited to tasks where rule-based answer verification is possible and does not naturally extend to real-world domains such as chemistry, healthcare, engineering, law, biology, business, and economics. Current practical workarounds use an additional LLM as a model-based verifier; however, this introduces issues such as reliance on a strong verifier LLM, susceptibility to reward hacking, and the practical burden of maintaining the verifier model in memory during training. To address this and extend DeepSeek-R1-Zero-style training to general reasoning domains, we propose a verifier-free method (VeriFree) that bypasses answer verification and instead uses RL to directly maximize the probability of generating the reference answer. We compare VeriFree with verifier-based methods and demonstrate that, in addition to its significant practical benefits and reduced compute requirements, VeriFree matches and even surpasses verifier-based methods on extensive evaluations across MMLU-Pro, GPQA, SuperGPQA, and math-related benchmarks. Moreover, we provide insights into this method from multiple perspectives: as an elegant integration of training both the policy and implicit verifier in a unified model, and as a variational optimization approach. Code is available at https://github.com/sail-sg/VeriFree.
Designing Cyclic Peptides via Harmonic SDE with Atom-Bond Modeling
May 27, 2025



Abstract:Cyclic peptides offer inherent advantages in pharmaceuticals. For example, cyclic peptides are more resistant to enzymatic hydrolysis compared to linear peptides and usually exhibit excellent stability and affinity. Although deep generative models have achieved great success in linear peptide design, several challenges prevent the development of computational methods for designing diverse types of cyclic peptides. These challenges include the scarcity of 3D structural data on target proteins and associated cyclic peptide ligands, the geometric constraints that cyclization imposes, and the involvement of non-canonical amino acids in cyclization. To address the above challenges, we introduce CpSDE, which consists of two key components: AtomSDE, a generative structure prediction model based on harmonic SDE, and ResRouter, a residue type predictor. Utilizing a routed sampling algorithm that alternates between these two models to iteratively update sequences and structures, CpSDE facilitates the generation of cyclic peptides. By employing explicit all-atom and bond modeling, CpSDE overcomes existing data limitations and is proficient in designing a wide variety of cyclic peptides. Our experimental results demonstrate that the cyclic peptides designed by our method exhibit reliable stability and affinity.
An All-Atom Generative Model for Designing Protein Complexes
Apr 17, 2025Abstract:Proteins typically exist in complexes, interacting with other proteins or biomolecules to perform their specific biological roles. Research on single-chain protein modeling has been extensively and deeply explored, with advancements seen in models like the series of ESM and AlphaFold. Despite these developments, the study and modeling of multi-chain proteins remain largely uncharted, though they are vital for understanding biological functions. Recognizing the importance of these interactions, we introduce APM (All-Atom Protein Generative Model), a model specifically designed for modeling multi-chain proteins. By integrating atom-level information and leveraging data on multi-chain proteins, APM is capable of precisely modeling inter-chain interactions and designing protein complexes with binding capabilities from scratch. It also performs folding and inverse-folding tasks for multi-chain proteins. Moreover, APM demonstrates versatility in downstream applications: it achieves enhanced performance through supervised fine-tuning (SFT) while also supporting zero-shot sampling in certain tasks, achieving state-of-the-art results. Code will be released at https://github.com/bytedance/apm.
Integrating Protein Dynamics into Structure-Based Drug Design via Full-Atom Stochastic Flows
Mar 06, 2025Abstract:The dynamic nature of proteins, influenced by ligand interactions, is essential for comprehending protein function and progressing drug discovery. Traditional structure-based drug design (SBDD) approaches typically target binding sites with rigid structures, limiting their practical application in drug development. While molecular dynamics simulation can theoretically capture all the biologically relevant conformations, the transition rate is dictated by the intrinsic energy barrier between them, making the sampling process computationally expensive. To overcome the aforementioned challenges, we propose to use generative modeling for SBDD considering conformational changes of protein pockets. We curate a dataset of apo and multiple holo states of protein-ligand complexes, simulated by molecular dynamics, and propose a full-atom flow model (and a stochastic version), named DynamicFlow, that learns to transform apo pockets and noisy ligands into holo pockets and corresponding 3D ligand molecules. Our method uncovers promising ligand molecules and corresponding holo conformations of pockets. Additionally, the resultant holo-like states provide superior inputs for traditional SBDD approaches, playing a significant role in practical drug discovery.
UniMatch: Universal Matching from Atom to Task for Few-Shot Drug Discovery
Feb 18, 2025Abstract:Drug discovery is crucial for identifying candidate drugs for various diseases.However, its low success rate often results in a scarcity of annotations, posing a few-shot learning problem. Existing methods primarily focus on single-scale features, overlooking the hierarchical molecular structures that determine different molecular properties. To address these issues, we introduce Universal Matching Networks (UniMatch), a dual matching framework that integrates explicit hierarchical molecular matching with implicit task-level matching via meta-learning, bridging multi-level molecular representations and task-level generalization. Specifically, our approach explicitly captures structural features across multiple levels, such as atoms, substructures, and molecules, via hierarchical pooling and matching, facilitating precise molecular representation and comparison. Additionally, we employ a meta-learning strategy for implicit task-level matching, allowing the model to capture shared patterns across tasks and quickly adapt to new ones. This unified matching framework ensures effective molecular alignment while leveraging shared meta-knowledge for fast adaptation. Our experimental results demonstrate that UniMatch outperforms state-of-the-art methods on the MoleculeNet and FS-Mol benchmarks, achieving improvements of 2.87% in AUROC and 6.52% in delta AUPRC. UniMatch also shows excellent generalization ability on the Meta-MolNet benchmark.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge