Yun Liu
Guangdong University of Finance, Guangzhou, China
Collision-Free Humanoid Traversal in Cluttered Indoor Scenes
Jan 23, 2026Abstract:We study the problem of collision-free humanoid traversal in cluttered indoor scenes, such as hurdling over objects scattered on the floor, crouching under low-hanging obstacles, or squeezing through narrow passages. To achieve this goal, the humanoid needs to map its perception of surrounding obstacles with diverse spatial layouts and geometries to the corresponding traversal skills. However, the lack of an effective representation that captures humanoid-obstacle relationships during collision avoidance makes directly learning such mappings difficult. We therefore propose Humanoid Potential Field (HumanoidPF), which encodes these relationships as collision-free motion directions, significantly facilitating RL-based traversal skill learning. We also find that HumanoidPF exhibits a surprisingly negligible sim-to-real gap as a perceptual representation. To further enable generalizable traversal skills through diverse and challenging cluttered indoor scenes, we further propose a hybrid scene generation method, incorporating crops of realistic 3D indoor scenes and procedurally synthesized obstacles. We successfully transfer our policy to the real world and develop a teleoperation system where users could command the humanoid to traverse in cluttered indoor scenes with just a single click. Extensive experiments are conducted in both simulation and the real world to validate the effectiveness of our method. Demos and code can be found in our website: https://axian12138.github.io/CAT/.
Evaluation of Large Language Models in Legal Applications: Challenges, Methods, and Future Directions
Jan 21, 2026Abstract:Large language models (LLMs) are being increasingly integrated into legal applications, including judicial decision support, legal practice assistance, and public-facing legal services. While LLMs show strong potential in handling legal knowledge and tasks, their deployment in real-world legal settings raises critical concerns beyond surface-level accuracy, involving the soundness of legal reasoning processes and trustworthy issues such as fairness and reliability. Systematic evaluation of LLM performance in legal tasks has therefore become essential for their responsible adoption. This survey identifies key challenges in evaluating LLMs for legal tasks grounded in real-world legal practice. We analyze the major difficulties involved in assessing LLM performance in the legal domain, including outcome correctness, reasoning reliability, and trustworthiness. Building on these challenges, we review and categorize existing evaluation methods and benchmarks according to their task design, datasets, and evaluation metrics. We further discuss the extent to which current approaches address these challenges, highlight their limitations, and outline future research directions toward more realistic, reliable, and legally grounded evaluation frameworks for LLMs in legal domains.
RealX3D: A Physically-Degraded 3D Benchmark for Multi-view Visual Restoration and Reconstruction
Dec 29, 2025Abstract:We introduce RealX3D, a real-capture benchmark for multi-view visual restoration and 3D reconstruction under diverse physical degradations. RealX3D groups corruptions into four families, including illumination, scattering, occlusion, and blurring, and captures each at multiple severity levels using a unified acquisition protocol that yields pixel-aligned LQ/GT views. Each scene includes high-resolution capture, RAW images, and dense laser scans, from which we derive world-scale meshes and metric depth. Benchmarking a broad range of optimization-based and feed-forward methods shows substantial degradation in reconstruction quality under physical corruptions, underscoring the fragility of current multi-view pipelines in real-world challenging environments.
Towards Interactive Intelligence for Digital Humans
Dec 15, 2025Abstract:We introduce Interactive Intelligence, a novel paradigm of digital human that is capable of personality-aligned expression, adaptive interaction, and self-evolution. To realize this, we present Mio (Multimodal Interactive Omni-Avatar), an end-to-end framework composed of five specialized modules: Thinker, Talker, Face Animator, Body Animator, and Renderer. This unified architecture integrates cognitive reasoning with real-time multimodal embodiment to enable fluid, consistent interaction. Furthermore, we establish a new benchmark to rigorously evaluate the capabilities of interactive intelligence. Extensive experiments demonstrate that our framework achieves superior performance compared to state-of-the-art methods across all evaluated dimensions. Together, these contributions move digital humans beyond superficial imitation toward intelligent interaction.
LexRel: Benchmarking Legal Relation Extraction for Chinese Civil Cases
Dec 14, 2025Abstract:Legal relations form a highly consequential analytical framework of civil law system, serving as a crucial foundation for resolving disputes and realizing values of the rule of law in judicial practice. However, legal relations in Chinese civil cases remain underexplored in the field of legal artificial intelligence (legal AI), largely due to the absence of comprehensive schemas. In this work, we firstly introduce a comprehensive schema, which contains a hierarchical taxonomy and definitions of arguments, for AI systems to capture legal relations in Chinese civil cases. Based on this schema, we then formulate legal relation extraction task and present LexRel, an expert-annotated benchmark for legal relation extraction in Chinese civil law. We use LexRel to evaluate state-of-the-art large language models (LLMs) on legal relation extractions, showing that current LLMs exhibit significant limitations in accurately identifying civil legal relations. Furthermore, we demonstrate that incorporating legal relations information leads to consistent performance gains on other downstream legal AI tasks.
UniLS: End-to-End Audio-Driven Avatars for Unified Listening and Speaking
Dec 10, 2025Abstract:Generating lifelike conversational avatars requires modeling not just isolated speakers, but the dynamic, reciprocal interaction of speaking and listening. However, modeling the listener is exceptionally challenging: direct audio-driven training fails, producing stiff, static listening motions. This failure stems from a fundamental imbalance: the speaker's motion is strongly driven by speech audio, while the listener's motion primarily follows an internal motion prior and is only loosely guided by external speech. This challenge has led most methods to focus on speak-only generation. The only prior attempt at joint generation relies on extra speaker's motion to produce the listener. This design is not end-to-end, thereby hindering the real-time applicability. To address this limitation, we present UniLS, the first end-to-end framework for generating unified speak-listen expressions, driven by only dual-track audio. Our method introduces a novel two-stage training paradigm. Stage 1 first learns the internal motion prior by training an audio-free autoregressive generator, capturing the spontaneous dynamics of natural facial motion. Stage 2 then introduces the dual-track audio, fine-tuning the generator to modulate the learned motion prior based on external speech cues. Extensive evaluations show UniLS achieves state-of-the-art speaking accuracy. More importantly, it delivers up to 44.1\% improvement in listening metrics, generating significantly more diverse and natural listening expressions. This effectively mitigates the stiffness problem and provides a practical, high-fidelity audio-driven solution for interactive digital humans.
Smartphone monitoring of smiling as a behavioral proxy of well-being in everyday life
Dec 10, 2025Abstract:Subjective well-being is a cornerstone of individual and societal health, yet its scientific measurement has traditionally relied on self-report methods prone to recall bias and high participant burden. This has left a gap in our understanding of well-being as it is expressed in everyday life. We hypothesized that candid smiles captured during natural smartphone interactions could serve as a scalable, objective behavioral correlate of positive affect. To test this, we analyzed 405,448 video clips passively recorded from 233 consented participants over one week. Using a deep learning model to quantify smile intensity, we identified distinct diurnal and daily patterns. Daily patterns of smile intensity across the week showed strong correlation with national survey data on happiness (r=0.92), and diurnal rhythms documented close correspondence with established results from the day reconstruction method (r=0.80). Higher daily mean smile intensity was significantly associated with more physical activity (Beta coefficient = 0.043, 95% CI [0.001, 0.085]) and greater light exposure (Beta coefficient = 0.038, [0.013, 0.063]), whereas no significant effects were found for smartphone use. These findings suggest that passive smartphone sensing could serve as a powerful, ecologically valid methodology for studying the dynamics of affective behavior and open the door to understanding this behavior at a population scale.
SFP: Real-World Scene Recovery Using Spatial and Frequency Priors
Dec 09, 2025



Abstract:Scene recovery serves as a critical task for various computer vision applications. Existing methods typically rely on a single prior, which is inherently insufficient to handle multiple degradations, or employ complex network architectures trained on synthetic data, which suffer from poor generalization for diverse real-world scenarios. In this paper, we propose Spatial and Frequency Priors (SFP) for real-world scene recovery. In the spatial domain, we observe that the inverse of the degraded image exhibits a projection along its spectral direction that resembles the scene transmission. Leveraging this spatial prior, the transmission map is estimated to recover the scene from scattering degradation. In the frequency domain, a mask is constructed for adaptive frequency enhancement, with two parameters estimated using our proposed novel priors. Specifically, one prior assumes that the mean intensity of the degraded image's direct current (DC) components across three channels in the frequency domain closely approximates that of each channel in the clear image. The second prior is based on the observation that, for clear images, the magnitude of low radial frequencies below 0.001 constitutes approximately 1% of the total spectrum. Finally, we design a weighted fusion strategy to integrate spatial-domain restoration, frequency-domain enhancement, and salient features from the input image, yielding the final recovered result. Extensive evaluations demonstrate the effectiveness and superiority of our proposed SFP for scene recovery under various degradation conditions.
Cardiac-CLIP: A Vision-Language Foundation Model for 3D Cardiac CT Images
Jul 29, 2025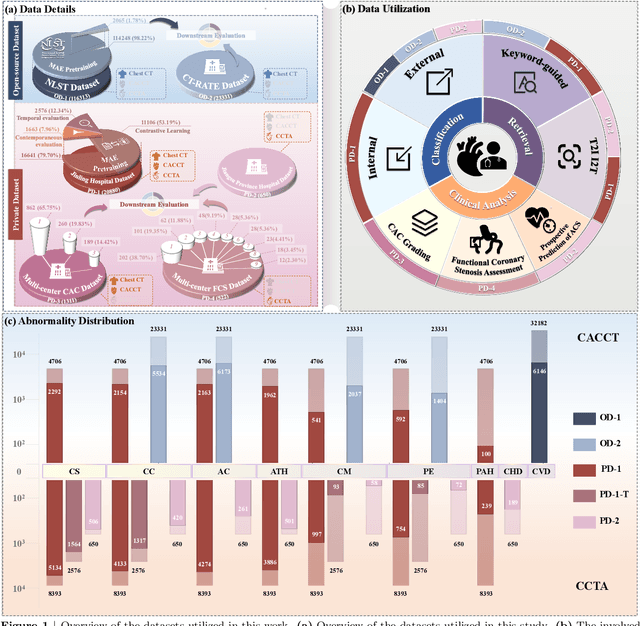
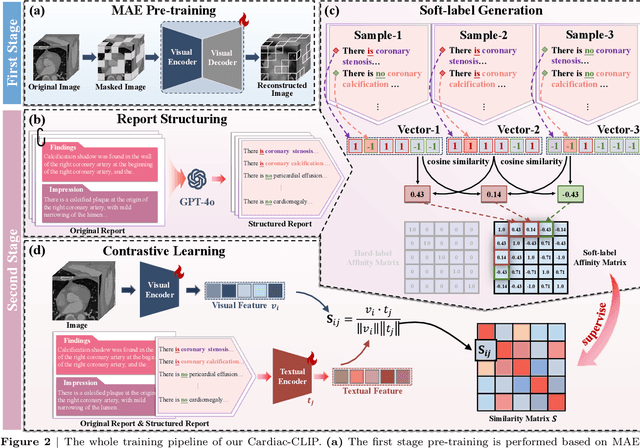
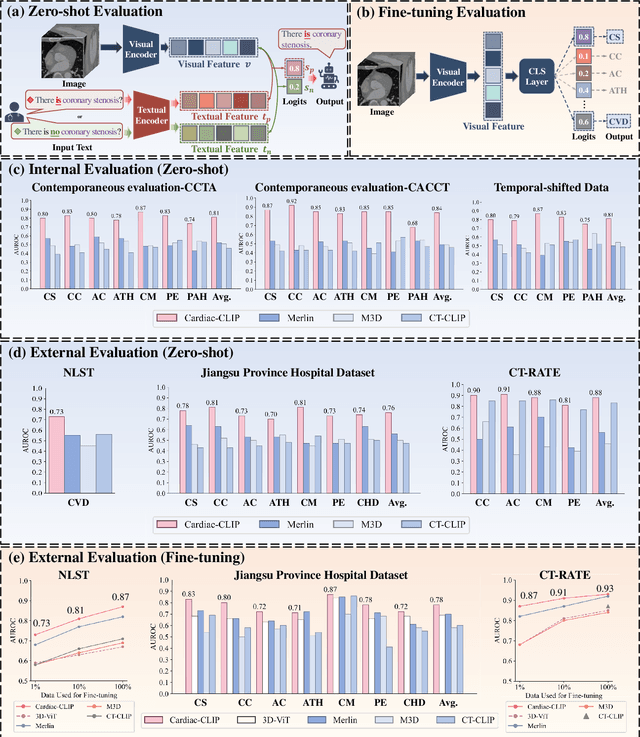
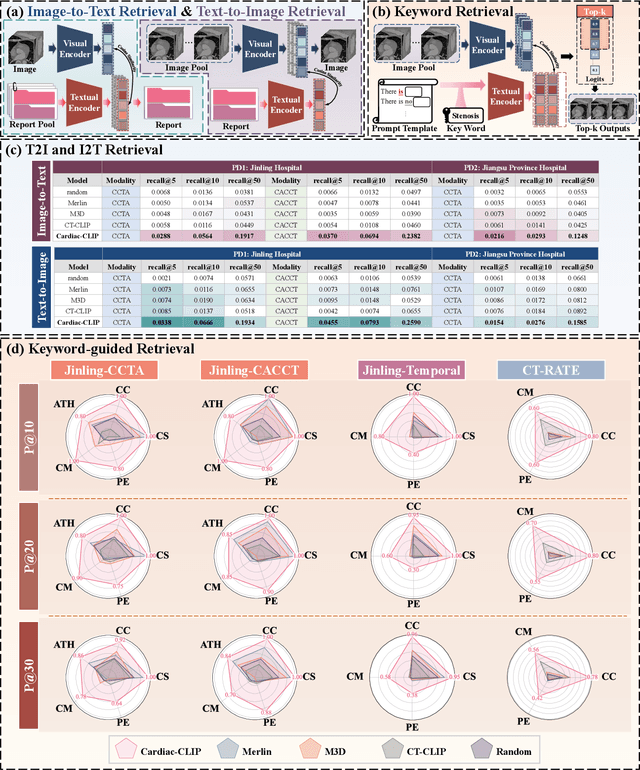
Abstract:Foundation models have demonstrated remarkable potential in medical domain. However, their application to complex cardiovascular diagnostics remains underexplored. In this paper, we present Cardiac-CLIP, a multi-modal foundation model designed for 3D cardiac CT images. Cardiac-CLIP is developed through a two-stage pre-training strategy. The first stage employs a 3D masked autoencoder (MAE) to perform self-supervised representation learning from large-scale unlabeled volumetric data, enabling the visual encoder to capture rich anatomical and contextual features. In the second stage, contrastive learning is introduced to align visual and textual representations, facilitating cross-modal understanding. To support the pre-training, we collect 16641 real clinical CT scans, supplemented by 114k publicly available data. Meanwhile, we standardize free-text radiology reports into unified templates and construct the pathology vectors according to diagnostic attributes, based on which the soft-label matrix is generated to supervise the contrastive learning process. On the other hand, to comprehensively evaluate the effectiveness of Cardiac-CLIP, we collect 6,722 real-clinical data from 12 independent institutions, along with the open-source data to construct the evaluation dataset. Specifically, Cardiac-CLIP is comprehensively evaluated across multiple tasks, including cardiovascular abnormality classification, information retrieval and clinical analysis. Experimental results demonstrate that Cardiac-CLIP achieves state-of-the-art performance across various downstream tasks in both internal and external data. Particularly, Cardiac-CLIP exhibits great effectiveness in supporting complex clinical tasks such as the prospective prediction of acute coronary syndrome, which is notoriously difficult in real-world scenarios.
A Comprehensive Survey on Video Scene Parsing:Advances, Challenges, and Prospects
Jun 16, 2025



Abstract:Video Scene Parsing (VSP) has emerged as a cornerstone in computer vision, facilitating the simultaneous segmentation, recognition, and tracking of diverse visual entities in dynamic scenes. In this survey, we present a holistic review of recent advances in VSP, covering a wide array of vision tasks, including Video Semantic Segmentation (VSS), Video Instance Segmentation (VIS), Video Panoptic Segmentation (VPS), as well as Video Tracking and Segmentation (VTS), and Open-Vocabulary Video Segmentation (OVVS). We systematically analyze the evolution from traditional hand-crafted features to modern deep learning paradigms -- spanning from fully convolutional networks to the latest transformer-based architectures -- and assess their effectiveness in capturing both local and global temporal contexts. Furthermore, our review critically discusses the technical challenges, ranging from maintaining temporal consistency to handling complex scene dynamics, and offers a comprehensive comparative study of datasets and evaluation metrics that have shaped current benchmarking standards. By distilling the key contributions and shortcomings of state-of-the-art methodologies, this survey highlights emerging trends and prospective research directions that promise to further elevate the robustness and adaptability of VSP in real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge