Maxwell A. Xu
SensorLM: Learning the Language of Wearable Sensors
Jun 10, 2025Abstract:We present SensorLM, a family of sensor-language foundation models that enable wearable sensor data understanding with natural language. Despite its pervasive nature, aligning and interpreting sensor data with language remains challenging due to the lack of paired, richly annotated sensor-text descriptions in uncurated, real-world wearable data. We introduce a hierarchical caption generation pipeline designed to capture statistical, structural, and semantic information from sensor data. This approach enabled the curation of the largest sensor-language dataset to date, comprising over 59.7 million hours of data from more than 103,000 people. Furthermore, SensorLM extends prominent multimodal pretraining architectures (e.g., CLIP, CoCa) and recovers them as specific variants within a generic architecture. Extensive experiments on real-world tasks in human activity analysis and healthcare verify the superior performance of SensorLM over state-of-the-art in zero-shot recognition, few-shot learning, and cross-modal retrieval. SensorLM also demonstrates intriguing capabilities including scaling behaviors, label efficiency, sensor captioning, and zero-shot generalization to unseen tasks.
RADAR: Benchmarking Language Models on Imperfect Tabular Data
Jun 09, 2025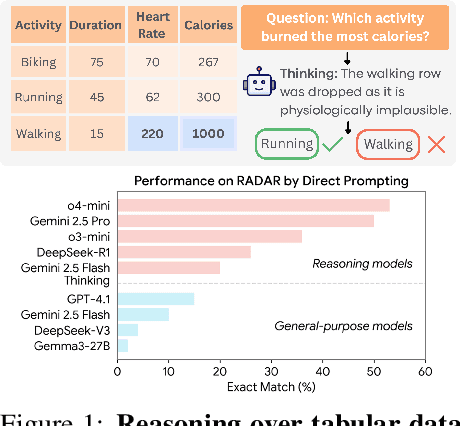
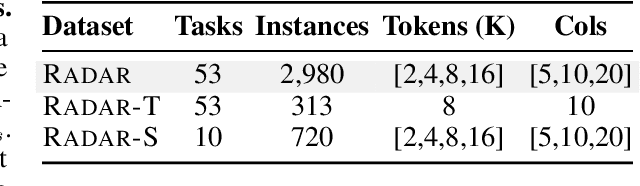
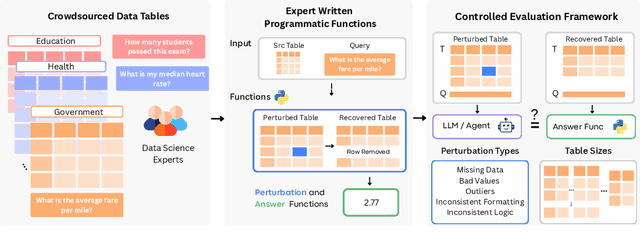
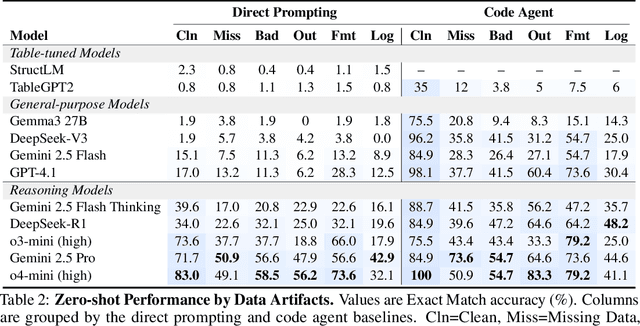
Abstract:Language models (LMs) are increasingly being deployed to perform autonomous data analyses. However, their data awareness -- the ability to recognize, reason over, and appropriately handle data artifacts such as missing values, outliers, and logical inconsistencies -- remains underexplored. These artifacts are especially common in real-world tabular data and, if mishandled, can significantly compromise the validity of analytical conclusions. To address this gap, we present RADAR, a benchmark for systematically evaluating data-aware reasoning on tabular data. We develop a framework to simulate data artifacts via programmatic perturbations to enable targeted evaluation of model behavior. RADAR comprises 2980 table query pairs, grounded in real-world data spanning 9 domains and 5 data artifact types. In addition to evaluating artifact handling, RADAR systematically varies table size to study how reasoning performance holds when increasing table size. Our evaluation reveals that, despite decent performance on tables without data artifacts, frontier models degrade significantly when data artifacts are introduced, exposing critical gaps in their capacity for robust, data-aware analysis. Designed to be flexible and extensible, RADAR supports diverse perturbation types and controllable table sizes, offering a valuable resource for advancing tabular reasoning.
LSM-2: Learning from Incomplete Wearable Sensor Data
Jun 05, 2025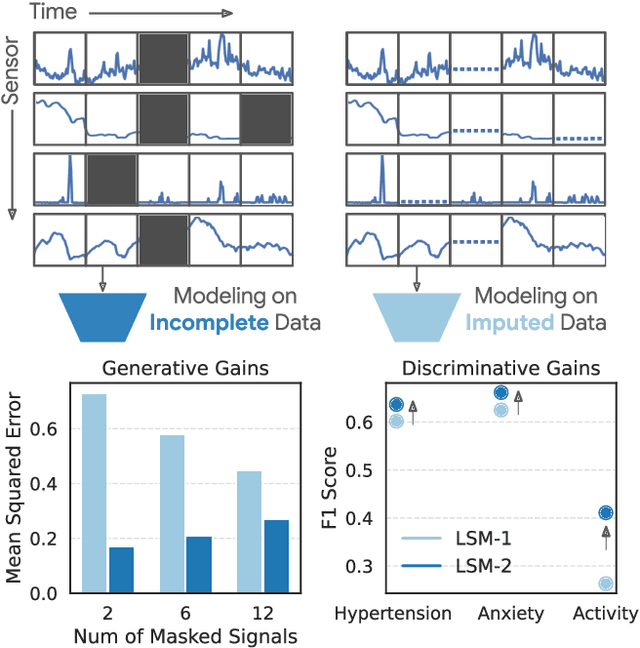
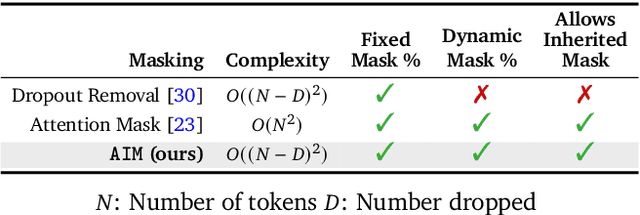
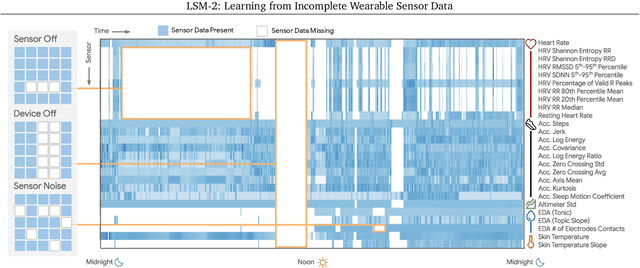
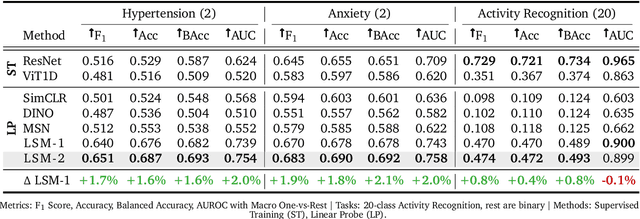
Abstract:Foundation models, a cornerstone of recent advancements in machine learning, have predominantly thrived on complete and well-structured data. Wearable sensor data frequently suffers from significant missingness, posing a substantial challenge for self-supervised learning (SSL) models that typically assume complete data inputs. This paper introduces the second generation of Large Sensor Model (LSM-2) with Adaptive and Inherited Masking (AIM), a novel SSL approach that learns robust representations directly from incomplete data without requiring explicit imputation. AIM's core novelty lies in its use of learnable mask tokens to model both existing ("inherited") and artificially introduced missingness, enabling it to robustly handle fragmented real-world data during inference. Pre-trained on an extensive dataset of 40M hours of day-long multimodal sensor data, our LSM-2 with AIM achieves the best performance across a diverse range of tasks, including classification, regression and generative modeling. Furthermore, LSM-2 with AIM exhibits superior scaling performance, and critically, maintains high performance even under targeted missingness scenarios, reflecting clinically coherent patterns, such as the diagnostic value of nighttime biosignals for hypertension prediction. This makes AIM a more reliable choice for real-world wearable data applications.
Pulse-PPG: An Open-Source Field-Trained PPG Foundation Model for Wearable Applications Across Lab and Field Settings
Feb 03, 2025Abstract:Photoplethysmography (PPG)-based foundation models are gaining traction due to the widespread use of PPG in biosignal monitoring and their potential to generalize across diverse health applications. In this paper, we introduce Pulse-PPG, the first open-source PPG foundation model trained exclusively on raw PPG data collected over a 100-day field study with 120 participants. Existing PPG foundation models are either open-source but trained on clinical data or closed-source, limiting their applicability in real-world settings. We evaluate Pulse-PPG across multiple datasets and downstream tasks, comparing its performance against a state-of-the-art foundation model trained on clinical data. Our results demonstrate that Pulse-PPG, trained on uncurated field data, exhibits superior generalization across clinical and mobile health applications in both lab and field settings. This suggests that exposure to real-world variability enables the model to learn fine-grained representations, making it more adaptable across tasks. Furthermore, pre-training on field data surprisingly outperforms its pre-training on clinical data in many tasks, reinforcing the importance of training on real-world, diverse datasets. To encourage further advancements in robust foundation models leveraging field data, we plan to release Pulse-PPG, providing researchers with a powerful resource for developing more generalizable PPG-based models.
PyPulse: A Python Library for Biosignal Imputation
Dec 09, 2024


Abstract:We introduce PyPulse, a Python package for imputation of biosignals in both clinical and wearable sensor settings. Missingness is commonplace in these settings and can arise from multiple causes, such as insecure sensor attachment or data transmission loss. PyPulse's framework provides a modular and extendable framework with high ease-of-use for a broad userbase, including non-machine-learning bioresearchers. Specifically, its new capabilities include using pre-trained imputation methods out-of-the-box on custom datasets, running the full workflow of training or testing a baseline method with a single line of code, and comparing baseline methods in an interactive visualization tool. We released PyPulse under the MIT License on Github and PyPI. The source code can be found at: https://github.com/rehg-lab/pulseimpute.
RelCon: Relative Contrastive Learning for a Motion Foundation Model for Wearable Data
Nov 27, 2024



Abstract:We present RelCon, a novel self-supervised \textit{Rel}ative \textit{Con}trastive learning approach that uses a learnable distance measure in combination with a softened contrastive loss for training an motion foundation model from wearable sensors. The learnable distance measure captures motif similarity and domain-specific semantic information such as rotation invariance. The learned distance provides a measurement of semantic similarity between a pair of accelerometer time-series segments, which is used to measure the distance between an anchor and various other sampled candidate segments. The self-supervised model is trained on 1 billion segments from 87,376 participants from a large wearables dataset. The model achieves strong performance across multiple downstream tasks, encompassing both classification and regression. To our knowledge, we are the first to show the generalizability of a self-supervised learning model with motion data from wearables across distinct evaluation tasks.
Towards Time Series Reasoning with LLMs
Sep 17, 2024Abstract:Multi-modal large language models (MLLMs) have enabled numerous advances in understanding and reasoning in domains like vision, but we have not yet seen this broad success for time-series. Although prior works on time-series MLLMs have shown promising performance in time-series forecasting, very few works show how an LLM could be used for time-series reasoning in natural language. We propose a novel multi-modal time-series LLM approach that learns generalizable information across various domains with powerful zero-shot performance. First, we train a lightweight time-series encoder on top of an LLM to directly extract time-series information. Then, we fine-tune our model with chain-of-thought augmented time-series tasks to encourage the model to generate reasoning paths. We show that our model learns a latent representation that reflects specific time-series features (e.g. slope, frequency), as well as outperforming GPT-4o on a set of zero-shot reasoning tasks on a variety of domains.
Temporally Multi-Scale Sparse Self-Attention for Physical Activity Data Imputation
Jun 27, 2024Abstract:Wearable sensors enable health researchers to continuously collect data pertaining to the physiological state of individuals in real-world settings. However, such data can be subject to extensive missingness due to a complex combination of factors. In this work, we study the problem of imputation of missing step count data, one of the most ubiquitous forms of wearable sensor data. We construct a novel and large scale data set consisting of a training set with over 3 million hourly step count observations and a test set with over 2.5 million hourly step count observations. We propose a domain knowledge-informed sparse self-attention model for this task that captures the temporal multi-scale nature of step-count data. We assess the performance of the model relative to baselines and conduct ablation studies to verify our specific model designs.
Retrieval-Based Reconstruction For Time-series Contrastive Learning
Nov 01, 2023Abstract:The success of self-supervised contrastive learning hinges on identifying positive data pairs that, when pushed together in embedding space, encode useful information for subsequent downstream tasks. However, in time-series, this is challenging because creating positive pairs via augmentations may break the original semantic meaning. We hypothesize that if we can retrieve information from one subsequence to successfully reconstruct another subsequence, then they should form a positive pair. Harnessing this intuition, we introduce our novel approach: REtrieval-BAsed Reconstruction (REBAR) contrastive learning. First, we utilize a convolutional cross-attention architecture to calculate the REBAR error between two different time-series. Then, through validation experiments, we show that the REBAR error is a predictor of mutual class membership, justifying its usage as a positive/negative labeler. Finally, once integrated into a contrastive learning framework, our REBAR method can learn an embedding that achieves state-of-the-art performance on downstream tasks across various modalities.
PulseImpute: A Novel Benchmark Task for Pulsative Physiological Signal Imputation
Dec 14, 2022Abstract:The promise of Mobile Health (mHealth) is the ability to use wearable sensors to monitor participant physiology at high frequencies during daily life to enable temporally-precise health interventions. However, a major challenge is frequent missing data. Despite a rich imputation literature, existing techniques are ineffective for the pulsative signals which comprise many mHealth applications, and a lack of available datasets has stymied progress. We address this gap with PulseImpute, the first large-scale pulsative signal imputation challenge which includes realistic mHealth missingness models, an extensive set of baselines, and clinically-relevant downstream tasks. Our baseline models include a novel transformer-based architecture designed to exploit the structure of pulsative signals. We hope that PulseImpute will enable the ML community to tackle this significant and challenging task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge