Jan Freyberg
Advancing Conversational Diagnostic AI with Multimodal Reasoning
May 06, 2025



Abstract:Large Language Models (LLMs) have demonstrated great potential for conducting diagnostic conversations but evaluation has been largely limited to language-only interactions, deviating from the real-world requirements of remote care delivery. Instant messaging platforms permit clinicians and patients to upload and discuss multimodal medical artifacts seamlessly in medical consultation, but the ability of LLMs to reason over such data while preserving other attributes of competent diagnostic conversation remains unknown. Here we advance the conversational diagnosis and management performance of the Articulate Medical Intelligence Explorer (AMIE) through a new capability to gather and interpret multimodal data, and reason about this precisely during consultations. Leveraging Gemini 2.0 Flash, our system implements a state-aware dialogue framework, where conversation flow is dynamically controlled by intermediate model outputs reflecting patient states and evolving diagnoses. Follow-up questions are strategically directed by uncertainty in such patient states, leading to a more structured multimodal history-taking process that emulates experienced clinicians. We compared AMIE to primary care physicians (PCPs) in a randomized, blinded, OSCE-style study of chat-based consultations with patient actors. We constructed 105 evaluation scenarios using artifacts like smartphone skin photos, ECGs, and PDFs of clinical documents across diverse conditions and demographics. Our rubric assessed multimodal capabilities and other clinically meaningful axes like history-taking, diagnostic accuracy, management reasoning, communication, and empathy. Specialist evaluation showed AMIE to be superior to PCPs on 7/9 multimodal and 29/32 non-multimodal axes (including diagnostic accuracy). The results show clear progress in multimodal conversational diagnostic AI, but real-world translation needs further research.
Accessible, At-Home Detection of Parkinson's Disease via Multi-task Video Analysis
Jun 21, 2024Abstract:Limited access to neurological care leads to missed diagnoses of Parkinson's disease (PD), leaving many individuals unidentified and untreated. We trained a novel neural network-based fusion architecture to detect Parkinson's disease (PD) by analyzing features extracted from webcam recordings of three tasks: finger tapping, facial expression (smiling), and speech (uttering a sentence containing all letters of the alphabet). Additionally, the model incorporated Monte Carlo Dropout to improve prediction accuracy by considering uncertainties. The study participants (n = 845, 272 with PD) were randomly split into three sets: 60% for training, 20% for model selection (hyper-parameter tuning), and 20% for final performance evaluation. The dataset consists of 1102 sessions, each session containing videos of all three tasks. Our proposed model achieved significantly better accuracy, area under the ROC curve (AUROC), and sensitivity at non-inferior specificity compared to any single-task model. Withholding uncertain predictions further boosted the performance, achieving 88.0% (95% CI: 87.7% - 88.4%) accuracy, 93.0% (92.8% - 93.2%) AUROC, 79.3% (78.4% - 80.2%) sensitivity, and 92.6% (92.3% - 92.8%) specificity, at the expense of not being able to predict for 2.3% (2.0% - 2.6%) data. Further analysis suggests that the trained model does not exhibit any detectable bias across sex and ethnic subgroups and is most effective for individuals aged between 50 and 80. This accessible, low-cost approach requiring only an internet-enabled device with a webcam and microphone paves the way for convenient PD screening at home, particularly in regions with limited access to clinical specialists.
Capabilities of Gemini Models in Medicine
May 01, 2024



Abstract:Excellence in a wide variety of medical applications poses considerable challenges for AI, requiring advanced reasoning, access to up-to-date medical knowledge and understanding of complex multimodal data. Gemini models, with strong general capabilities in multimodal and long-context reasoning, offer exciting possibilities in medicine. Building on these core strengths of Gemini, we introduce Med-Gemini, a family of highly capable multimodal models that are specialized in medicine with the ability to seamlessly use web search, and that can be efficiently tailored to novel modalities using custom encoders. We evaluate Med-Gemini on 14 medical benchmarks, establishing new state-of-the-art (SoTA) performance on 10 of them, and surpass the GPT-4 model family on every benchmark where a direct comparison is viable, often by a wide margin. On the popular MedQA (USMLE) benchmark, our best-performing Med-Gemini model achieves SoTA performance of 91.1% accuracy, using a novel uncertainty-guided search strategy. On 7 multimodal benchmarks including NEJM Image Challenges and MMMU (health & medicine), Med-Gemini improves over GPT-4V by an average relative margin of 44.5%. We demonstrate the effectiveness of Med-Gemini's long-context capabilities through SoTA performance on a needle-in-a-haystack retrieval task from long de-identified health records and medical video question answering, surpassing prior bespoke methods using only in-context learning. Finally, Med-Gemini's performance suggests real-world utility by surpassing human experts on tasks such as medical text summarization, alongside demonstrations of promising potential for multimodal medical dialogue, medical research and education. Taken together, our results offer compelling evidence for Med-Gemini's potential, although further rigorous evaluation will be crucial before real-world deployment in this safety-critical domain.
Closing the AI generalization gap by adjusting for dermatology condition distribution differences across clinical settings
Feb 23, 2024Abstract:Recently, there has been great progress in the ability of artificial intelligence (AI) algorithms to classify dermatological conditions from clinical photographs. However, little is known about the robustness of these algorithms in real-world settings where several factors can lead to a loss of generalizability. Understanding and overcoming these limitations will permit the development of generalizable AI that can aid in the diagnosis of skin conditions across a variety of clinical settings. In this retrospective study, we demonstrate that differences in skin condition distribution, rather than in demographics or image capture mode are the main source of errors when an AI algorithm is evaluated on data from a previously unseen source. We demonstrate a series of steps to close this generalization gap, requiring progressively more information about the new source, ranging from the condition distribution to training data enriched for data less frequently seen during training. Our results also suggest comparable performance from end-to-end fine tuning versus fine tuning solely the classification layer on top of a frozen embedding model. Our approach can inform the adaptation of AI algorithms to new settings, based on the information and resources available.
MINT: A wrapper to make multi-modal and multi-image AI models interactive
Jan 22, 2024Abstract:During the diagnostic process, doctors incorporate multimodal information including imaging and the medical history - and similarly medical AI development has increasingly become multimodal. In this paper we tackle a more subtle challenge: doctors take a targeted medical history to obtain only the most pertinent pieces of information; how do we enable AI to do the same? We develop a wrapper method named MINT (Make your model INTeractive) that automatically determines what pieces of information are most valuable at each step, and ask for only the most useful information. We demonstrate the efficacy of MINT wrapping a skin disease prediction model, where multiple images and a set of optional answers to $25$ standard metadata questions (i.e., structured medical history) are used by a multi-modal deep network to provide a differential diagnosis. We show that MINT can identify whether metadata inputs are needed and if so, which question to ask next. We also demonstrate that when collecting multiple images, MINT can identify if an additional image would be beneficial, and if so, which type of image to capture. We showed that MINT reduces the number of metadata and image inputs needed by 82% and 36.2% respectively, while maintaining predictive performance. Using real-world AI dermatology system data, we show that needing fewer inputs can retain users that may otherwise fail to complete the system submission and drop off without a diagnosis. Qualitative examples show MINT can closely mimic the step-by-step decision making process of a clinical workflow and how this is different for straight forward cases versus more difficult, ambiguous cases. Finally we demonstrate how MINT is robust to different underlying multi-model classifiers and can be easily adapted to user requirements without significant model re-training.
Towards Conversational Diagnostic AI
Jan 11, 2024Abstract:At the heart of medicine lies the physician-patient dialogue, where skillful history-taking paves the way for accurate diagnosis, effective management, and enduring trust. Artificial Intelligence (AI) systems capable of diagnostic dialogue could increase accessibility, consistency, and quality of care. However, approximating clinicians' expertise is an outstanding grand challenge. Here, we introduce AMIE (Articulate Medical Intelligence Explorer), a Large Language Model (LLM) based AI system optimized for diagnostic dialogue. AMIE uses a novel self-play based simulated environment with automated feedback mechanisms for scaling learning across diverse disease conditions, specialties, and contexts. We designed a framework for evaluating clinically-meaningful axes of performance including history-taking, diagnostic accuracy, management reasoning, communication skills, and empathy. We compared AMIE's performance to that of primary care physicians (PCPs) in a randomized, double-blind crossover study of text-based consultations with validated patient actors in the style of an Objective Structured Clinical Examination (OSCE). The study included 149 case scenarios from clinical providers in Canada, the UK, and India, 20 PCPs for comparison with AMIE, and evaluations by specialist physicians and patient actors. AMIE demonstrated greater diagnostic accuracy and superior performance on 28 of 32 axes according to specialist physicians and 24 of 26 axes according to patient actors. Our research has several limitations and should be interpreted with appropriate caution. Clinicians were limited to unfamiliar synchronous text-chat which permits large-scale LLM-patient interactions but is not representative of usual clinical practice. While further research is required before AMIE could be translated to real-world settings, the results represent a milestone towards conversational diagnostic AI.
Evaluating AI systems under uncertain ground truth: a case study in dermatology
Jul 05, 2023



Abstract:For safety, AI systems in health undergo thorough evaluations before deployment, validating their predictions against a ground truth that is assumed certain. However, this is actually not the case and the ground truth may be uncertain. Unfortunately, this is largely ignored in standard evaluation of AI models but can have severe consequences such as overestimating the future performance. To avoid this, we measure the effects of ground truth uncertainty, which we assume decomposes into two main components: annotation uncertainty which stems from the lack of reliable annotations, and inherent uncertainty due to limited observational information. This ground truth uncertainty is ignored when estimating the ground truth by deterministically aggregating annotations, e.g., by majority voting or averaging. In contrast, we propose a framework where aggregation is done using a statistical model. Specifically, we frame aggregation of annotations as posterior inference of so-called plausibilities, representing distributions over classes in a classification setting, subject to a hyper-parameter encoding annotator reliability. Based on this model, we propose a metric for measuring annotation uncertainty and provide uncertainty-adjusted metrics for performance evaluation. We present a case study applying our framework to skin condition classification from images where annotations are provided in the form of differential diagnoses. The deterministic adjudication process called inverse rank normalization (IRN) from previous work ignores ground truth uncertainty in evaluation. Instead, we present two alternative statistical models: a probabilistic version of IRN and a Plackett-Luce-based model. We find that a large portion of the dataset exhibits significant ground truth uncertainty and standard IRN-based evaluation severely over-estimates performance without providing uncertainty estimates.
Interactive Concept Bottleneck Models
Dec 26, 2022



Abstract:Concept bottleneck models (CBMs) (Koh et al. 2020) are interpretable neural networks that first predict labels for human-interpretable concepts relevant to the prediction task, and then predict the final label based on the concept label predictions.We extend CBMs to interactive prediction settings where the model can query a human collaborator for the label to some concepts. We develop an interaction policy that, at prediction time, chooses which concepts to request a label for so as to maximally improve the final prediction. We demonstrate thata simple policy combining concept prediction uncertainty and influence of the concept on the final prediction achieves strong performance and outperforms a static approach proposed in Koh et al. (2020) as well as active feature acquisition methods proposed in the literature. We show that the interactiveCBM can achieve accuracy gains of 5-10% with only 5 interactions over competitive baselines on the Caltech-UCSDBirds, CheXpert and OAI datasets.
Detecting and Preventing Shortcut Learning for Fair Medical AI using Shortcut Testing (ShorT)
Jul 21, 2022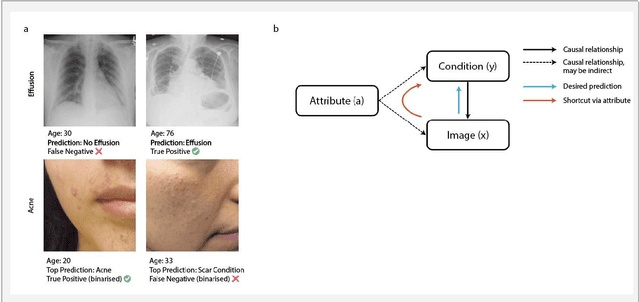
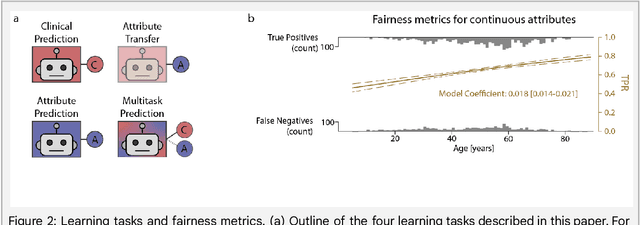
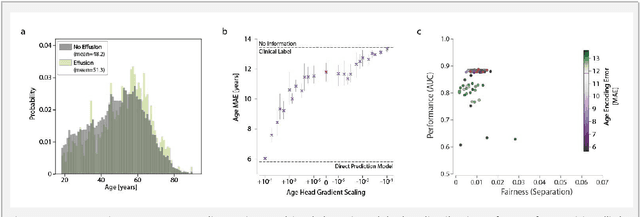
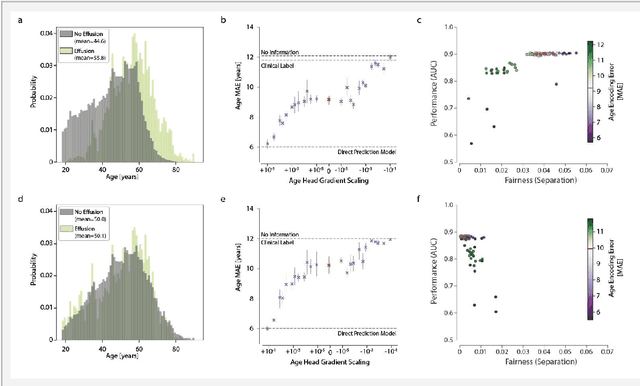
Abstract:Machine learning (ML) holds great promise for improving healthcare, but it is critical to ensure that its use will not propagate or amplify health disparities. An important step is to characterize the (un)fairness of ML models - their tendency to perform differently across subgroups of the population - and to understand its underlying mechanisms. One potential driver of algorithmic unfairness, shortcut learning, arises when ML models base predictions on improper correlations in the training data. However, diagnosing this phenomenon is difficult, especially when sensitive attributes are causally linked with disease. Using multi-task learning, we propose the first method to assess and mitigate shortcut learning as a part of the fairness assessment of clinical ML systems, and demonstrate its application to clinical tasks in radiology and dermatology. Finally, our approach reveals instances when shortcutting is not responsible for unfairness, highlighting the need for a holistic approach to fairness mitigation in medical AI.
Robust and Efficient Medical Imaging with Self-Supervision
May 19, 2022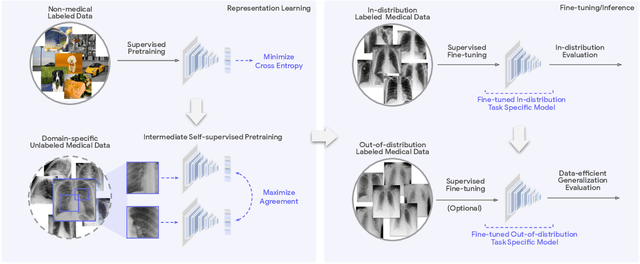
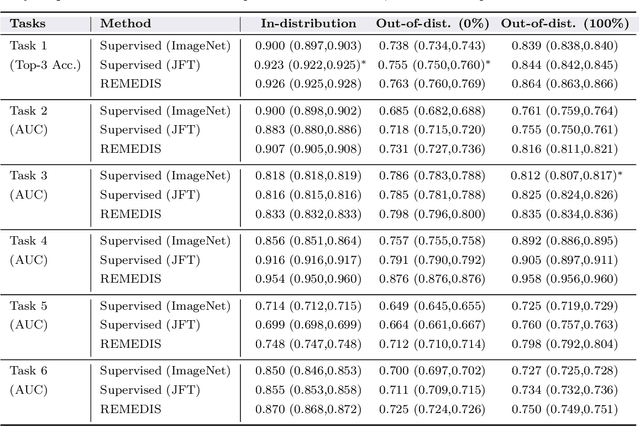
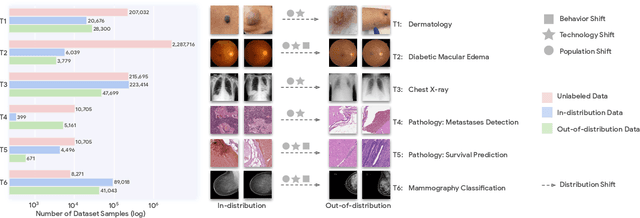
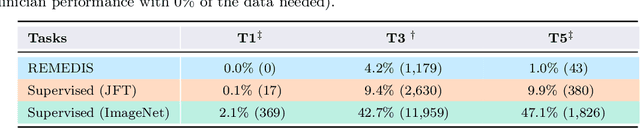
Abstract:Recent progress in Medical Artificial Intelligence (AI) has delivered systems that can reach clinical expert level performance. However, such systems tend to demonstrate sub-optimal "out-of-distribution" performance when evaluated in clinical settings different from the training environment. A common mitigation strategy is to develop separate systems for each clinical setting using site-specific data [1]. However, this quickly becomes impractical as medical data is time-consuming to acquire and expensive to annotate [2]. Thus, the problem of "data-efficient generalization" presents an ongoing difficulty for Medical AI development. Although progress in representation learning shows promise, their benefits have not been rigorously studied, specifically for out-of-distribution settings. To meet these challenges, we present REMEDIS, a unified representation learning strategy to improve robustness and data-efficiency of medical imaging AI. REMEDIS uses a generic combination of large-scale supervised transfer learning with self-supervised learning and requires little task-specific customization. We study a diverse range of medical imaging tasks and simulate three realistic application scenarios using retrospective data. REMEDIS exhibits significantly improved in-distribution performance with up to 11.5% relative improvement in diagnostic accuracy over a strong supervised baseline. More importantly, our strategy leads to strong data-efficient generalization of medical imaging AI, matching strong supervised baselines using between 1% to 33% of retraining data across tasks. These results suggest that REMEDIS can significantly accelerate the life-cycle of medical imaging AI development thereby presenting an important step forward for medical imaging AI to deliver broad impact.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge