A. Ali Heydari
Towards a Science of Scaling Agent Systems
Dec 17, 2025Abstract:Agents, language model-based systems that are capable of reasoning, planning, and acting are becoming the dominant paradigm for real-world AI applications. Despite this widespread adoption, the principles that determine their performance remain underexplored. We address this by deriving quantitative scaling principles for agent systems. We first formalize a definition for agentic evaluation and characterize scaling laws as the interplay between agent quantity, coordination structure, model capability, and task properties. We evaluate this across four benchmarks: Finance-Agent, BrowseComp-Plus, PlanCraft, and Workbench. With five canonical agent architectures (Single-Agent and four Multi-Agent Systems: Independent, Centralized, Decentralized, Hybrid), instantiated across three LLM families, we perform a controlled evaluation spanning 180 configurations. We derive a predictive model using coordination metrics, that achieves cross-validated R^2=0.524, enabling prediction on unseen task domains. We identify three effects: (1) a tool-coordination trade-off: under fixed computational budgets, tool-heavy tasks suffer disproportionately from multi-agent overhead. (2) a capability saturation: coordination yields diminishing or negative returns once single-agent baselines exceed ~45%. (3) topology-dependent error amplification: independent agents amplify errors 17.2x, while centralized coordination contains this to 4.4x. Centralized coordination improves performance by 80.8% on parallelizable tasks, while decentralized coordination excels on web navigation (+9.2% vs. +0.2%). Yet for sequential reasoning tasks, every multi-agent variants degraded performance by 39-70%. The framework predicts the optimal coordination strategy for 87% of held-out configurations. Out-of-sample validation on GPT-5.2, achieves MAE=0.071 and confirms four of five scaling principles generalize to unseen frontier models.
SensorLM: Learning the Language of Wearable Sensors
Jun 10, 2025Abstract:We present SensorLM, a family of sensor-language foundation models that enable wearable sensor data understanding with natural language. Despite its pervasive nature, aligning and interpreting sensor data with language remains challenging due to the lack of paired, richly annotated sensor-text descriptions in uncurated, real-world wearable data. We introduce a hierarchical caption generation pipeline designed to capture statistical, structural, and semantic information from sensor data. This approach enabled the curation of the largest sensor-language dataset to date, comprising over 59.7 million hours of data from more than 103,000 people. Furthermore, SensorLM extends prominent multimodal pretraining architectures (e.g., CLIP, CoCa) and recovers them as specific variants within a generic architecture. Extensive experiments on real-world tasks in human activity analysis and healthcare verify the superior performance of SensorLM over state-of-the-art in zero-shot recognition, few-shot learning, and cross-modal retrieval. SensorLM also demonstrates intriguing capabilities including scaling behaviors, label efficiency, sensor captioning, and zero-shot generalization to unseen tasks.
RADAR: Benchmarking Language Models on Imperfect Tabular Data
Jun 09, 2025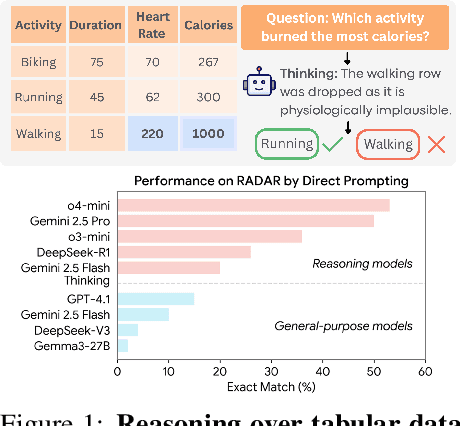
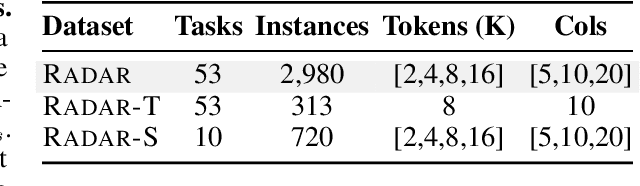
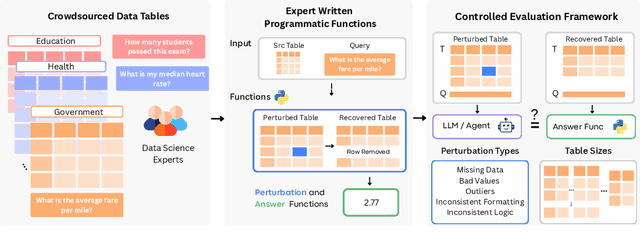
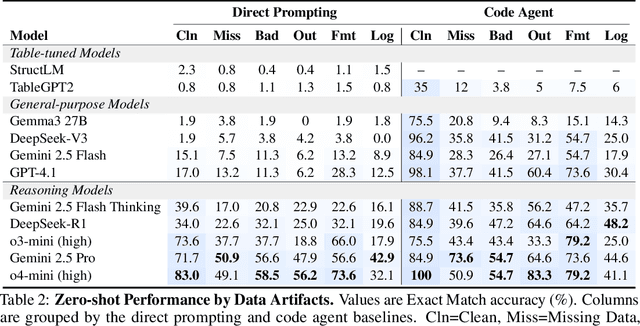
Abstract:Language models (LMs) are increasingly being deployed to perform autonomous data analyses. However, their data awareness -- the ability to recognize, reason over, and appropriately handle data artifacts such as missing values, outliers, and logical inconsistencies -- remains underexplored. These artifacts are especially common in real-world tabular data and, if mishandled, can significantly compromise the validity of analytical conclusions. To address this gap, we present RADAR, a benchmark for systematically evaluating data-aware reasoning on tabular data. We develop a framework to simulate data artifacts via programmatic perturbations to enable targeted evaluation of model behavior. RADAR comprises 2980 table query pairs, grounded in real-world data spanning 9 domains and 5 data artifact types. In addition to evaluating artifact handling, RADAR systematically varies table size to study how reasoning performance holds when increasing table size. Our evaluation reveals that, despite decent performance on tables without data artifacts, frontier models degrade significantly when data artifacts are introduced, exposing critical gaps in their capacity for robust, data-aware analysis. Designed to be flexible and extensible, RADAR supports diverse perturbation types and controllable table sizes, offering a valuable resource for advancing tabular reasoning.
LSM-2: Learning from Incomplete Wearable Sensor Data
Jun 05, 2025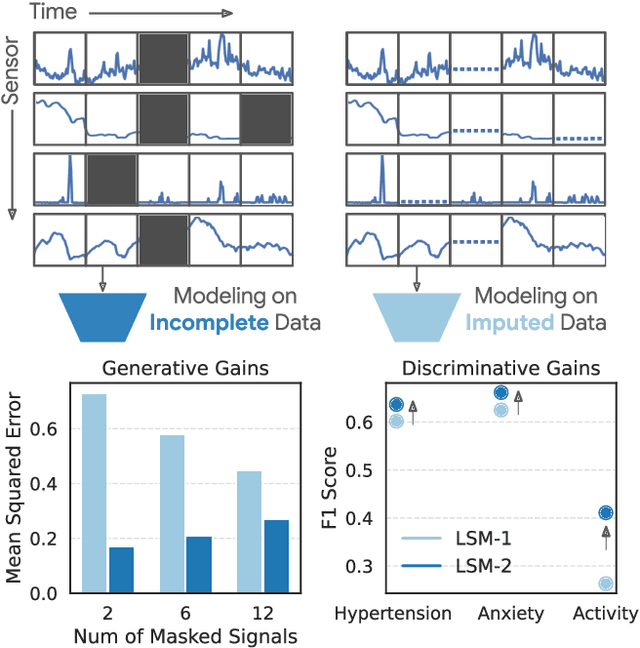
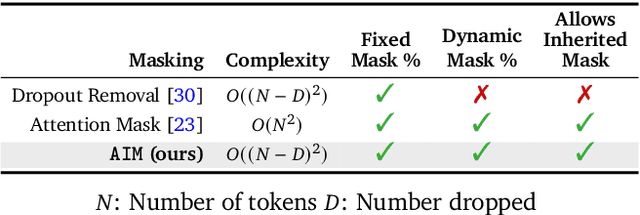
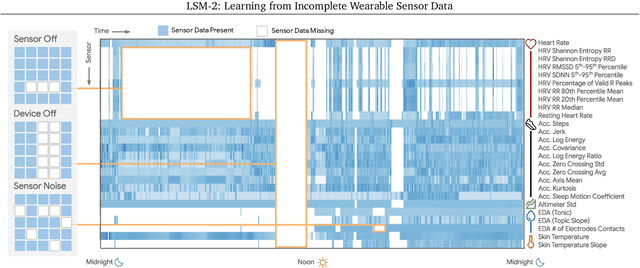
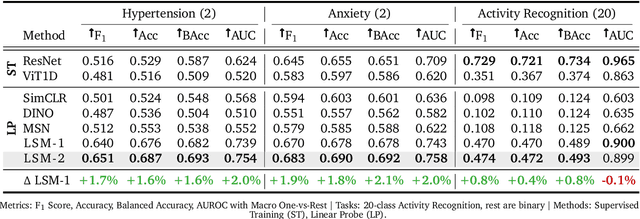
Abstract:Foundation models, a cornerstone of recent advancements in machine learning, have predominantly thrived on complete and well-structured data. Wearable sensor data frequently suffers from significant missingness, posing a substantial challenge for self-supervised learning (SSL) models that typically assume complete data inputs. This paper introduces the second generation of Large Sensor Model (LSM-2) with Adaptive and Inherited Masking (AIM), a novel SSL approach that learns robust representations directly from incomplete data without requiring explicit imputation. AIM's core novelty lies in its use of learnable mask tokens to model both existing ("inherited") and artificially introduced missingness, enabling it to robustly handle fragmented real-world data during inference. Pre-trained on an extensive dataset of 40M hours of day-long multimodal sensor data, our LSM-2 with AIM achieves the best performance across a diverse range of tasks, including classification, regression and generative modeling. Furthermore, LSM-2 with AIM exhibits superior scaling performance, and critically, maintains high performance even under targeted missingness scenarios, reflecting clinically coherent patterns, such as the diagnostic value of nighttime biosignals for hypertension prediction. This makes AIM a more reliable choice for real-world wearable data applications.
Insulin Resistance Prediction From Wearables and Routine Blood Biomarkers
Apr 30, 2025Abstract:Insulin resistance, a precursor to type 2 diabetes, is characterized by impaired insulin action in tissues. Current methods for measuring insulin resistance, while effective, are expensive, inaccessible, not widely available and hinder opportunities for early intervention. In this study, we remotely recruited the largest dataset to date across the US to study insulin resistance (N=1,165 participants, with median BMI=28 kg/m2, age=45 years, HbA1c=5.4%), incorporating wearable device time series data and blood biomarkers, including the ground-truth measure of insulin resistance, homeostatic model assessment for insulin resistance (HOMA-IR). We developed deep neural network models to predict insulin resistance based on readily available digital and blood biomarkers. Our results show that our models can predict insulin resistance by combining both wearable data and readily available blood biomarkers better than either of the two data sources separately (R2=0.5, auROC=0.80, Sensitivity=76%, and specificity 84%). The model showed 93% sensitivity and 95% adjusted specificity in obese and sedentary participants, a subpopulation most vulnerable to developing type 2 diabetes and who could benefit most from early intervention. Rigorous evaluation of model performance, including interpretability, and robustness, facilitates generalizability across larger cohorts, which is demonstrated by reproducing the prediction performance on an independent validation cohort (N=72 participants). Additionally, we demonstrated how the predicted insulin resistance can be integrated into a large language model agent to help understand and contextualize HOMA-IR values, facilitating interpretation and safe personalized recommendations. This work offers the potential for early detection of people at risk of type 2 diabetes and thereby facilitate earlier implementation of preventative strategies.
A Scalable Framework for Evaluating Health Language Models
Apr 01, 2025Abstract:Large language models (LLMs) have emerged as powerful tools for analyzing complex datasets. Recent studies demonstrate their potential to generate useful, personalized responses when provided with patient-specific health information that encompasses lifestyle, biomarkers, and context. As LLM-driven health applications are increasingly adopted, rigorous and efficient one-sided evaluation methodologies are crucial to ensure response quality across multiple dimensions, including accuracy, personalization and safety. Current evaluation practices for open-ended text responses heavily rely on human experts. This approach introduces human factors and is often cost-prohibitive, labor-intensive, and hinders scalability, especially in complex domains like healthcare where response assessment necessitates domain expertise and considers multifaceted patient data. In this work, we introduce Adaptive Precise Boolean rubrics: an evaluation framework that streamlines human and automated evaluation of open-ended questions by identifying gaps in model responses using a minimal set of targeted rubrics questions. Our approach is based on recent work in more general evaluation settings that contrasts a smaller set of complex evaluation targets with a larger set of more precise, granular targets answerable with simple boolean responses. We validate this approach in metabolic health, a domain encompassing diabetes, cardiovascular disease, and obesity. Our results demonstrate that Adaptive Precise Boolean rubrics yield higher inter-rater agreement among expert and non-expert human evaluators, and in automated assessments, compared to traditional Likert scales, while requiring approximately half the evaluation time of Likert-based methods. This enhanced efficiency, particularly in automated evaluation and non-expert contributions, paves the way for more extensive and cost-effective evaluation of LLMs in health.
Lifestyle-Informed Personalized Blood Biomarker Prediction via Novel Representation Learning
Jul 09, 2024



Abstract:Blood biomarkers are an essential tool for healthcare providers to diagnose, monitor, and treat a wide range of medical conditions. Current reference values and recommended ranges often rely on population-level statistics, which may not adequately account for the influence of inter-individual variability driven by factors such as lifestyle and genetics. In this work, we introduce a novel framework for predicting future blood biomarker values and define personalized references through learned representations from lifestyle data (physical activity and sleep) and blood biomarkers. Our proposed method learns a similarity-based embedding space that captures the complex relationship between biomarkers and lifestyle factors. Using the UK Biobank (257K participants), our results show that our deep-learned embeddings outperform traditional and current state-of-the-art representation learning techniques in predicting clinical diagnosis. Using a subset of UK Biobank of 6440 participants who have follow-up visits, we validate that the inclusion of these embeddings and lifestyle factors directly in blood biomarker models improves the prediction of future lab values from a single lab visit. This personalized modeling approach provides a foundation for developing more accurate risk stratification tools and tailoring preventative care strategies. In clinical settings, this translates to the potential for earlier disease detection, more timely interventions, and ultimately, a shift towards personalized healthcare.
No Pairs Left Behind: Improving Metric Learning with Regularized Triplet Objective
Oct 18, 2022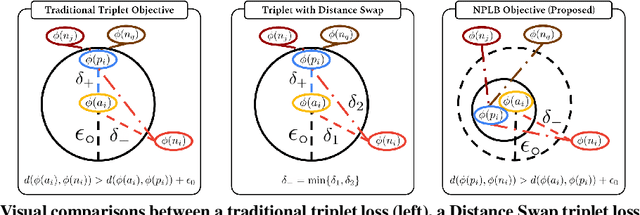

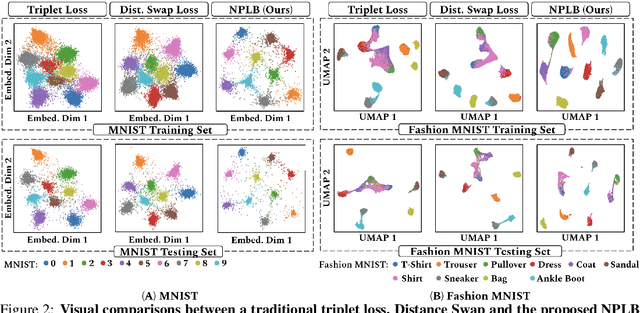
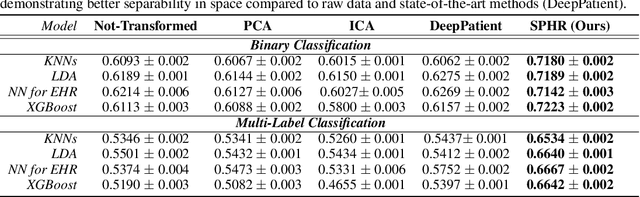
Abstract:We propose a novel formulation of the triplet objective function that improves metric learning without additional sample mining or overhead costs. Our approach aims to explicitly regularize the distance between the positive and negative samples in a triplet with respect to the anchor-negative distance. As an initial validation, we show that our method (called No Pairs Left Behind [NPLB]) improves upon the traditional and current state-of-the-art triplet objective formulations on standard benchmark datasets. To show the effectiveness and potentials of NPLB on real-world complex data, we evaluate our approach on a large-scale healthcare dataset (UK Biobank), demonstrating that the embeddings learned by our model significantly outperform all other current representations on tested downstream tasks. Additionally, we provide a new model-agnostic single-time health risk definition that, when used in tandem with the learned representations, achieves the most accurate prediction of subjects' future health complications. Our results indicate that NPLB is a simple, yet effective framework for improving existing deep metric learning models, showcasing the potential implications of metric learning in more complex applications, especially in the biological and healthcare domains.
SoftAdapt: Techniques for Adaptive Loss Weighting of Neural Networks with Multi-Part Loss Functions
Dec 27, 2019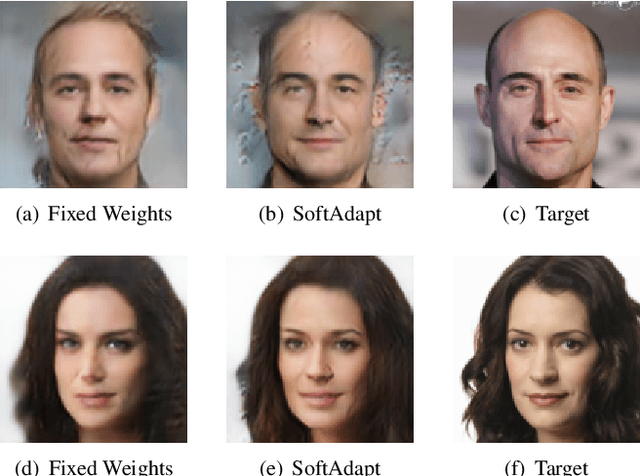
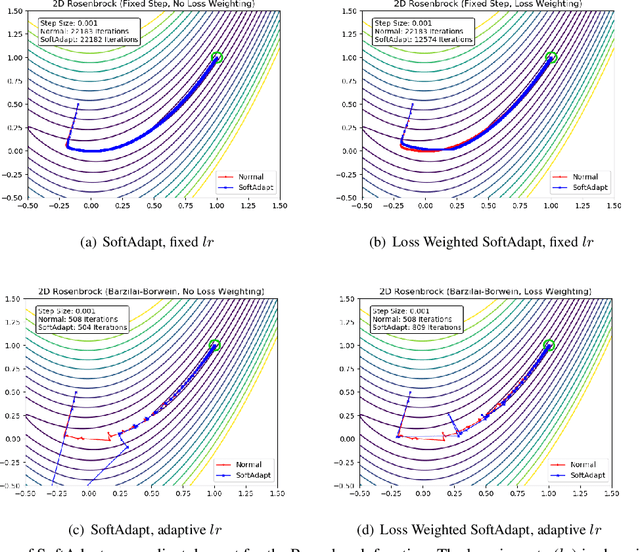
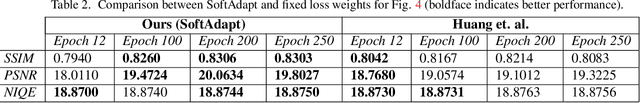

Abstract:Adaptive loss function formulation is an active area of research and has gained a great deal of popularity in recent years, following the success of deep learning. However, existing frameworks of adaptive loss functions often suffer from slow convergence and poor choice of weights for the loss components. Traditionally, the elements of a multi-part loss function are weighted equally or their weights are determined through heuristic approaches that yield near-optimal (or sub-optimal) results. To address this problem, we propose a family of methods, called SoftAdapt, that dynamically change function weights for multi-part loss functions based on live performance statistics of the component losses. SoftAdapt is mathematically intuitive, computationally efficient and straightforward to implement. In this paper, we present the mathematical formulation and pseudocode for SoftAdapt, along with results from applying our methods to image reconstruction (Sparse Autoencoders) and synthetic data generation (Introspective Variational Autoencoders).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge