Yu Shi
3DGAUnet: 3D generative adversarial networks with a 3D U-Net based generator to achieve the accurate and effective synthesis of clinical tumor image data for pancreatic cancer
Nov 27, 2023Abstract:Pancreatic ductal adenocarcinoma (PDAC) presents a critical global health challenge, and early detection is crucial for improving the 5-year survival rate. Recent medical imaging and computational algorithm advances offer potential solutions for early diagnosis. Deep learning, particularly in the form of convolutional neural networks (CNNs), has demonstrated success in medical image analysis tasks, including classification and segmentation. However, the limited availability of clinical data for training purposes continues to provide a significant obstacle. Data augmentation, generative adversarial networks (GANs), and cross-validation are potential techniques to address this limitation and improve model performance, but effective solutions are still rare for 3D PDAC, where contrast is especially poor owing to the high heterogeneity in both tumor and background tissues. In this study, we developed a new GAN-based model, named 3DGAUnet, for generating realistic 3D CT images of PDAC tumors and pancreatic tissue, which can generate the interslice connection data that the existing 2D CT image synthesis models lack. Our innovation is to develop a 3D U-Net architecture for the generator to improve shape and texture learning for PDAC tumors and pancreatic tissue. Our approach offers a promising path to tackle the urgent requirement for creative and synergistic methods to combat PDAC. The development of this GAN-based model has the potential to alleviate data scarcity issues, elevate the quality of synthesized data, and thereby facilitate the progression of deep learning models to enhance the accuracy and early detection of PDAC tumors, which could profoundly impact patient outcomes. Furthermore, this model has the potential to be adapted to other types of solid tumors, hence making significant contributions to the field of medical imaging in terms of image processing models.
Robust Representation Learning for Unreliable Partial Label Learning
Aug 31, 2023



Abstract:Partial Label Learning (PLL) is a type of weakly supervised learning where each training instance is assigned a set of candidate labels, but only one label is the ground-truth. However, this idealistic assumption may not always hold due to potential annotation inaccuracies, meaning the ground-truth may not be present in the candidate label set. This is known as Unreliable Partial Label Learning (UPLL) that introduces an additional complexity due to the inherent unreliability and ambiguity of partial labels, often resulting in a sub-optimal performance with existing methods. To address this challenge, we propose the Unreliability-Robust Representation Learning framework (URRL) that leverages unreliability-robust contrastive learning to help the model fortify against unreliable partial labels effectively. Concurrently, we propose a dual strategy that combines KNN-based candidate label set correction and consistency-regularization-based label disambiguation to refine label quality and enhance the ability of representation learning within the URRL framework. Extensive experiments demonstrate that the proposed method outperforms state-of-the-art PLL methods on various datasets with diverse degrees of unreliability and ambiguity. Furthermore, we provide a theoretical analysis of our approach from the perspective of the expectation maximization (EM) algorithm. Upon acceptance, we pledge to make the code publicly accessible.
SLPT: Selective Labeling Meets Prompt Tuning on Label-Limited Lesion Segmentation
Aug 09, 2023


Abstract:Medical image analysis using deep learning is often challenged by limited labeled data and high annotation costs. Fine-tuning the entire network in label-limited scenarios can lead to overfitting and suboptimal performance. Recently, prompt tuning has emerged as a more promising technique that introduces a few additional tunable parameters as prompts to a task-agnostic pre-trained model, and updates only these parameters using supervision from limited labeled data while keeping the pre-trained model unchanged. However, previous work has overlooked the importance of selective labeling in downstream tasks, which aims to select the most valuable downstream samples for annotation to achieve the best performance with minimum annotation cost. To address this, we propose a framework that combines selective labeling with prompt tuning (SLPT) to boost performance in limited labels. Specifically, we introduce a feature-aware prompt updater to guide prompt tuning and a TandEm Selective LAbeling (TESLA) strategy. TESLA includes unsupervised diversity selection and supervised selection using prompt-based uncertainty. In addition, we propose a diversified visual prompt tuning strategy to provide multi-prompt-based discrepant predictions for TESLA. We evaluate our method on liver tumor segmentation and achieve state-of-the-art performance, outperforming traditional fine-tuning with only 6% of tunable parameters, also achieving 94% of full-data performance by labeling only 5% of the data.
Improved Prognostic Prediction of Pancreatic Cancer Using Multi-Phase CT by Integrating Neural Distance and Texture-Aware Transformer
Aug 01, 2023



Abstract:Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal cancer in which the tumor-vascular involvement greatly affects the resectability and, thus, overall survival of patients. However, current prognostic prediction methods fail to explicitly and accurately investigate relationships between the tumor and nearby important vessels. This paper proposes a novel learnable neural distance that describes the precise relationship between the tumor and vessels in CT images of different patients, adopting it as a major feature for prognosis prediction. Besides, different from existing models that used CNNs or LSTMs to exploit tumor enhancement patterns on dynamic contrast-enhanced CT imaging, we improved the extraction of dynamic tumor-related texture features in multi-phase contrast-enhanced CT by fusing local and global features using CNN and transformer modules, further enhancing the features extracted across multi-phase CT images. We extensively evaluated and compared the proposed method with existing methods in the multi-center (n=4) dataset with 1,070 patients with PDAC, and statistical analysis confirmed its clinical effectiveness in the external test set consisting of three centers. The developed risk marker was the strongest predictor of overall survival among preoperative factors and it has the potential to be combined with established clinical factors to select patients at higher risk who might benefit from neoadjuvant therapy.
Liver Tumor Screening and Diagnosis in CT with Pixel-Lesion-Patient Network
Jul 17, 2023Abstract:Liver tumor segmentation and classification are important tasks in computer aided diagnosis. We aim to address three problems: liver tumor screening and preliminary diagnosis in non-contrast computed tomography (CT), and differential diagnosis in dynamic contrast-enhanced CT. A novel framework named Pixel-Lesion-pAtient Network (PLAN) is proposed. It uses a mask transformer to jointly segment and classify each lesion with improved anchor queries and a foreground-enhanced sampling loss. It also has an image-wise classifier to effectively aggregate global information and predict patient-level diagnosis. A large-scale multi-phase dataset is collected containing 939 tumor patients and 810 normal subjects. 4010 tumor instances of eight types are extensively annotated. On the non-contrast tumor screening task, PLAN achieves 95% and 96% in patient-level sensitivity and specificity. On contrast-enhanced CT, our lesion-level detection precision, recall, and classification accuracy are 92%, 89%, and 86%, outperforming widely used CNN and transformers for lesion segmentation. We also conduct a reader study on a holdout set of 250 cases. PLAN is on par with a senior human radiologist, showing the clinical significance of our results.
Towards Predicting Equilibrium Distributions for Molecular Systems with Deep Learning
Jun 08, 2023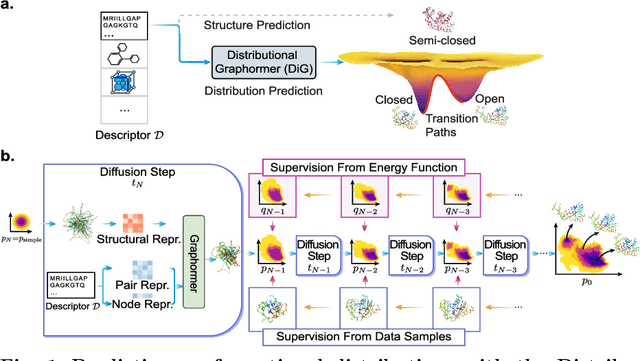
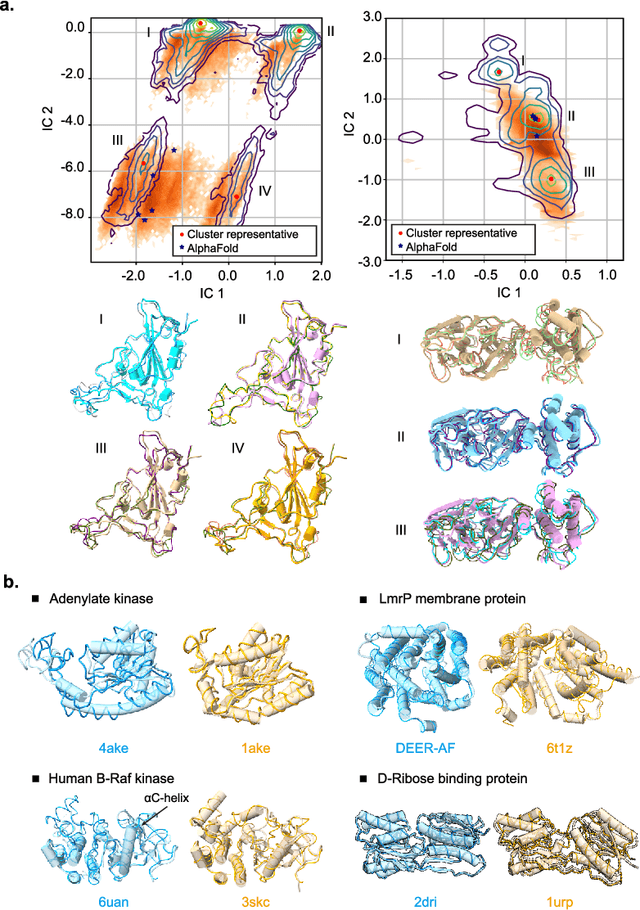
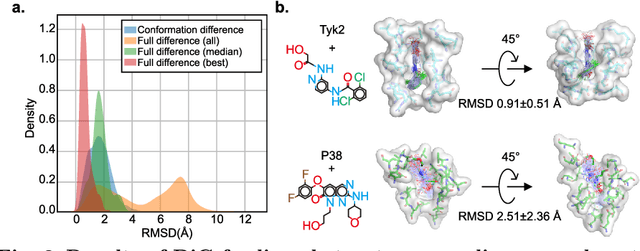
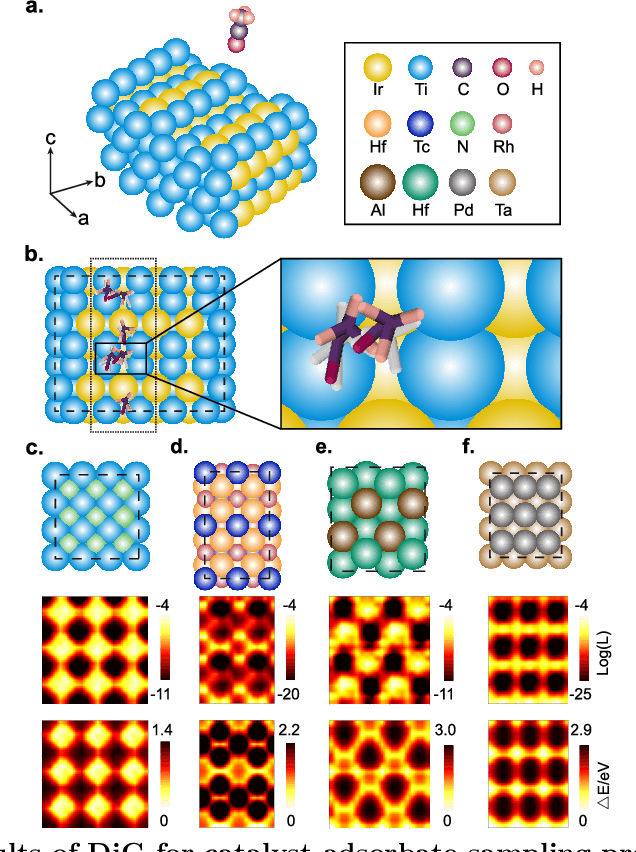
Abstract:Advances in deep learning have greatly improved structure prediction of molecules. However, many macroscopic observations that are important for real-world applications are not functions of a single molecular structure, but rather determined from the equilibrium distribution of structures. Traditional methods for obtaining these distributions, such as molecular dynamics simulation, are computationally expensive and often intractable. In this paper, we introduce a novel deep learning framework, called Distributional Graphormer (DiG), in an attempt to predict the equilibrium distribution of molecular systems. Inspired by the annealing process in thermodynamics, DiG employs deep neural networks to transform a simple distribution towards the equilibrium distribution, conditioned on a descriptor of a molecular system, such as a chemical graph or a protein sequence. This framework enables efficient generation of diverse conformations and provides estimations of state densities. We demonstrate the performance of DiG on several molecular tasks, including protein conformation sampling, ligand structure sampling, catalyst-adsorbate sampling, and property-guided structure generation. DiG presents a significant advancement in methodology for statistically understanding molecular systems, opening up new research opportunities in molecular science.
Breaking the Curse of Quality Saturation with User-Centric Ranking
May 24, 2023



Abstract:A key puzzle in search, ads, and recommendation is that the ranking model can only utilize a small portion of the vastly available user interaction data. As a result, increasing data volume, model size, or computation FLOPs will quickly suffer from diminishing returns. We examined this problem and found that one of the root causes may lie in the so-called ``item-centric'' formulation, which has an unbounded vocabulary and thus uncontrolled model complexity. To mitigate quality saturation, we introduce an alternative formulation named ``user-centric ranking'', which is based on a transposed view of the dyadic user-item interaction data. We show that this formulation has a promising scaling property, enabling us to train better-converged models on substantially larger data sets.
i-Code V2: An Autoregressive Generation Framework over Vision, Language, and Speech Data
May 21, 2023



Abstract:The convergence of text, visual, and audio data is a key step towards human-like artificial intelligence, however the current Vision-Language-Speech landscape is dominated by encoder-only models which lack generative abilities. We propose closing this gap with i-Code V2, the first model capable of generating natural language from any combination of Vision, Language, and Speech data. i-Code V2 is an integrative system that leverages state-of-the-art single-modality encoders, combining their outputs with a new modality-fusing encoder in order to flexibly project combinations of modalities into a shared representational space. Next, language tokens are generated from these representations via an autoregressive decoder. The whole framework is pretrained end-to-end on a large collection of dual- and single-modality datasets using a novel text completion objective that can be generalized across arbitrary combinations of modalities. i-Code V2 matches or outperforms state-of-the-art single- and dual-modality baselines on 7 multimodal tasks, demonstrating the power of generative multimodal pretraining across a diversity of tasks and signals.
Devil is in the Queries: Advancing Mask Transformers for Real-world Medical Image Segmentation and Out-of-Distribution Localization
Apr 01, 2023



Abstract:Real-world medical image segmentation has tremendous long-tailed complexity of objects, among which tail conditions correlate with relatively rare diseases and are clinically significant. A trustworthy medical AI algorithm should demonstrate its effectiveness on tail conditions to avoid clinically dangerous damage in these out-of-distribution (OOD) cases. In this paper, we adopt the concept of object queries in Mask Transformers to formulate semantic segmentation as a soft cluster assignment. The queries fit the feature-level cluster centers of inliers during training. Therefore, when performing inference on a medical image in real-world scenarios, the similarity between pixels and the queries detects and localizes OOD regions. We term this OOD localization as MaxQuery. Furthermore, the foregrounds of real-world medical images, whether OOD objects or inliers, are lesions. The difference between them is less than that between the foreground and background, possibly misleading the object queries to focus redundantly on the background. Thus, we propose a query-distribution (QD) loss to enforce clear boundaries between segmentation targets and other regions at the query level, improving the inlier segmentation and OOD indication. Our proposed framework is tested on two real-world segmentation tasks, i.e., segmentation of pancreatic and liver tumors, outperforming previous state-of-the-art algorithms by an average of 7.39% on AUROC, 14.69% on AUPR, and 13.79% on FPR95 for OOD localization. On the other hand, our framework improves the performance of inlier segmentation by an average of 5.27% DSC when compared with the leading baseline nnUNet.
Code-Switching Text Generation and Injection in Mandarin-English ASR
Mar 20, 2023



Abstract:Code-switching speech refers to a means of expression by mixing two or more languages within a single utterance. Automatic Speech Recognition (ASR) with End-to-End (E2E) modeling for such speech can be a challenging task due to the lack of data. In this study, we investigate text generation and injection for improving the performance of an industry commonly-used streaming model, Transformer-Transducer (T-T), in Mandarin-English code-switching speech recognition. We first propose a strategy to generate code-switching text data and then investigate injecting generated text into T-T model explicitly by Text-To-Speech (TTS) conversion or implicitly by tying speech and text latent spaces. Experimental results on the T-T model trained with a dataset containing 1,800 hours of real Mandarin-English code-switched speech show that our approaches to inject generated code-switching text significantly boost the performance of T-T models, i.e., 16% relative Token-based Error Rate (TER) reduction averaged on three evaluation sets, and the approach of tying speech and text latent spaces is superior to that of TTS conversion on the evaluation set which contains more homogeneous data with the training set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge