A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020Zhaohan Xiong, Qing Xia, Zhiqiang Hu, Ning Huang, Cheng Bian, Yefeng Zheng, Sulaiman Vesal, Nishant Ravikumar, Andreas Maier, Xin Yang, Pheng-Ann Heng, Dong Ni, Caizi Li, Qianqian Tong, Weixin Si, Elodie Puybareau, Younes Khoudli, Thierry Geraud, Chen Chen, Wenjia Bai, Daniel Rueckert, Lingchao Xu, Xiahai Zhuang, Xinzhe Luo, Shuman Jia, Maxime Sermesant, Yashu Liu, Kuanquan Wang, Davide Borra, Alessandro Masci, Cristiana Corsi, Coen de Vente, Mitko Veta, Rashed Karim, Chandrakanth Jayachandran Preetha, Sandy Engelhardt, Menyun Qiao, Yuanyuan Wang, Qian Tao, Marta Nunez-Garcia, Oscar Camara, Nicolo Savioli, Pablo Lamata, Jichao Zhao
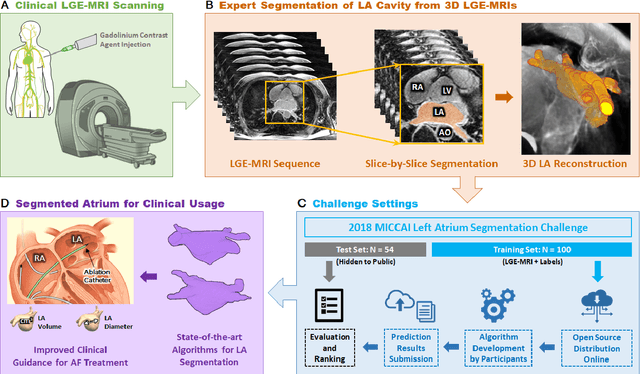
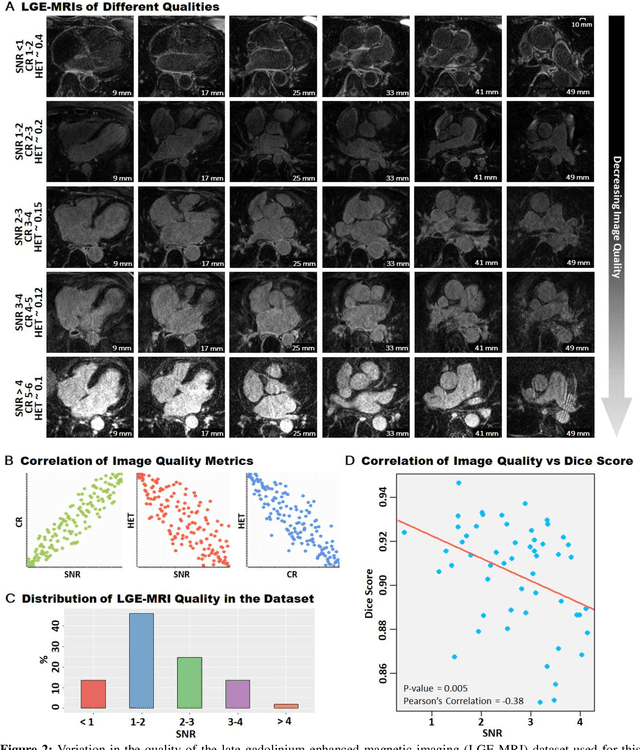
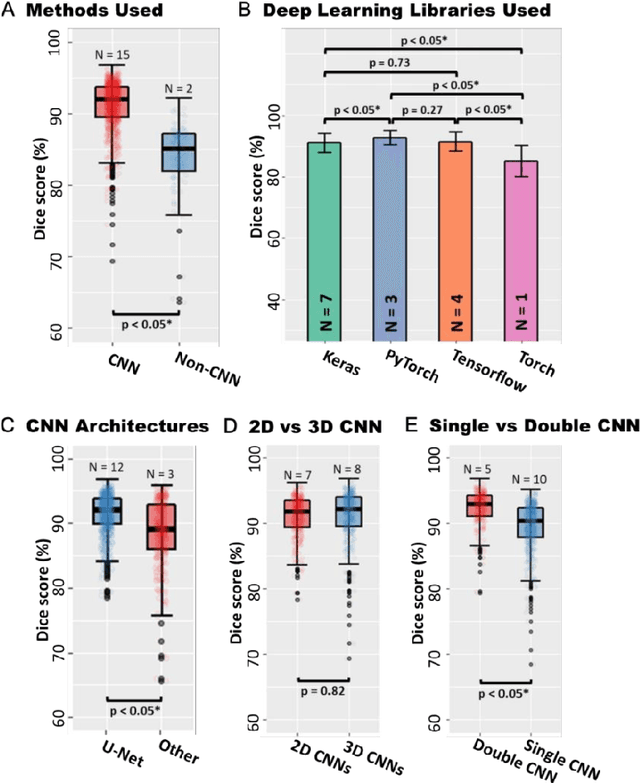
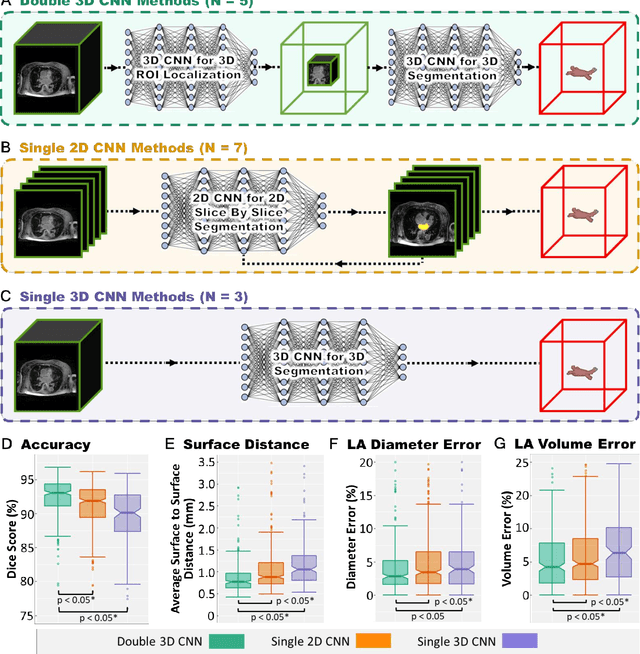
Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
Quantifying Graft Detachment after Descemet's Membrane Endothelial Keratoplasty with Deep Convolutional Neural Networks
Apr 24, 2020Friso G. Heslinga, Mark Alberti, Josien P. W. Pluim, Javier Cabrerizo, Mitko Veta
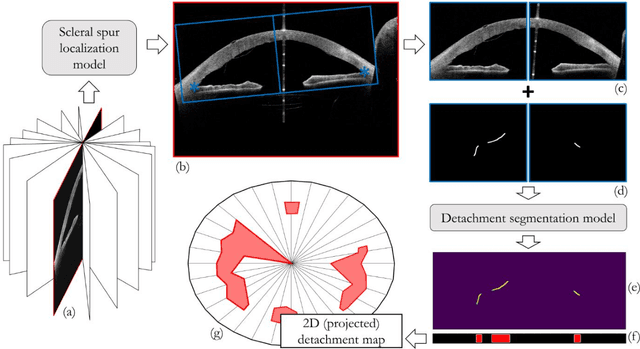
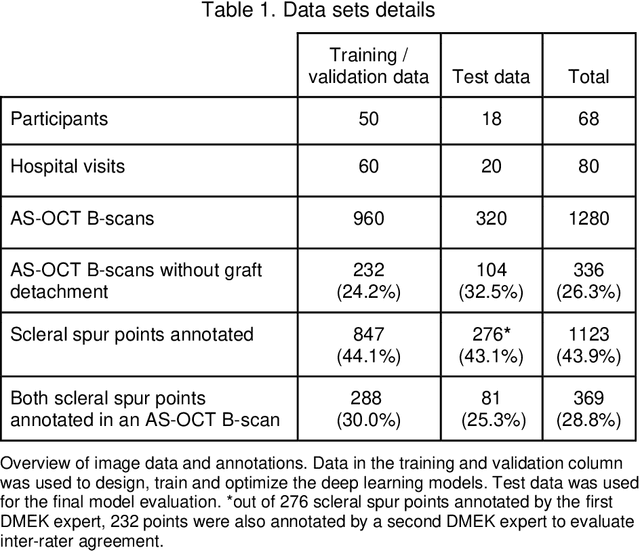
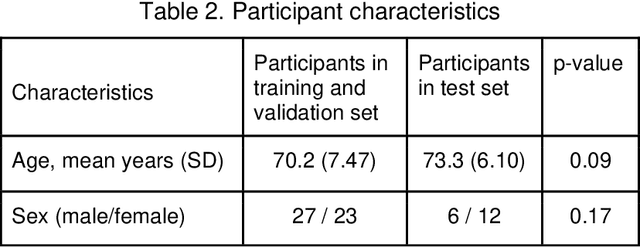
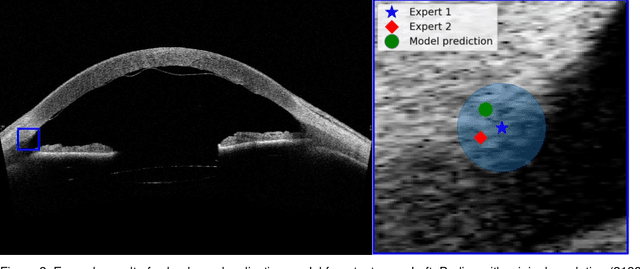
Purpose: We developed a method to automatically locate and quantify graft detachment after Descemet's Membrane Endothelial Keratoplasty (DMEK) in Anterior Segment Optical Coherence Tomography (AS-OCT) scans. Methods: 1280 AS-OCT B-scans were annotated by a DMEK expert. Using the annotations, a deep learning pipeline was developed to localize scleral spur, center the AS-OCT B-scans and segment the detached graft sections. Detachment segmentation model performance was evaluated per B-scan by comparing (1) length of detachment and (2) horizontal projection of the detached sections with the expert annotations. Horizontal projections were used to construct graft detachment maps. All final evaluations were done on a test set that was set apart during training of the models. A second DMEK expert annotated the test set to determine inter-rater performance. Results: Mean scleral spur localization error was 0.155 mm, whereas the inter-rater difference was 0.090 mm. The estimated graft detachment lengths were in 69% of the cases within a 10-pixel (~150{\mu}m) difference from the ground truth (77% for the second DMEK expert). Dice scores for the horizontal projections of all B-scans with detachments were 0.896 and 0.880 for our model and the second DMEK expert respectively. Conclusion: Our deep learning model can be used to automatically and instantly localize graft detachment in AS-OCT B-scans. Horizontal detachment projections can be determined with the same accuracy as a human DMEK expert, allowing for the construction of accurate graft detachment maps. Translational Relevance: Automated localization and quantification of graft detachment can support DMEK research and standardize clinical decision making.
Roto-Translation Equivariant Convolutional Networks: Application to Histopathology Image Analysis
Feb 20, 2020Maxime W. Lafarge, Erik J. Bekkers, Josien P. W. Pluim, Remco Duits, Mitko Veta
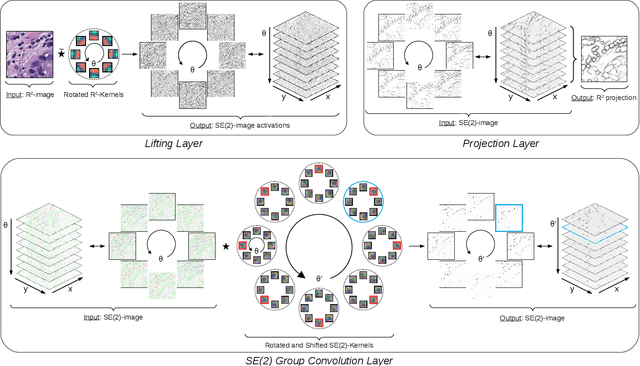
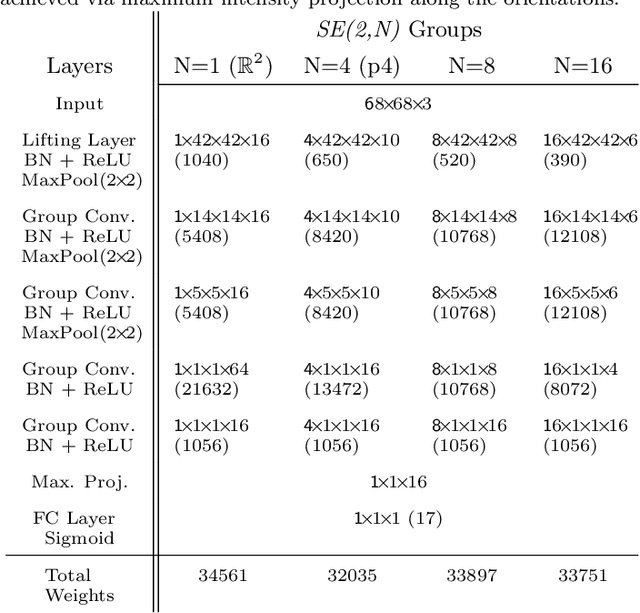
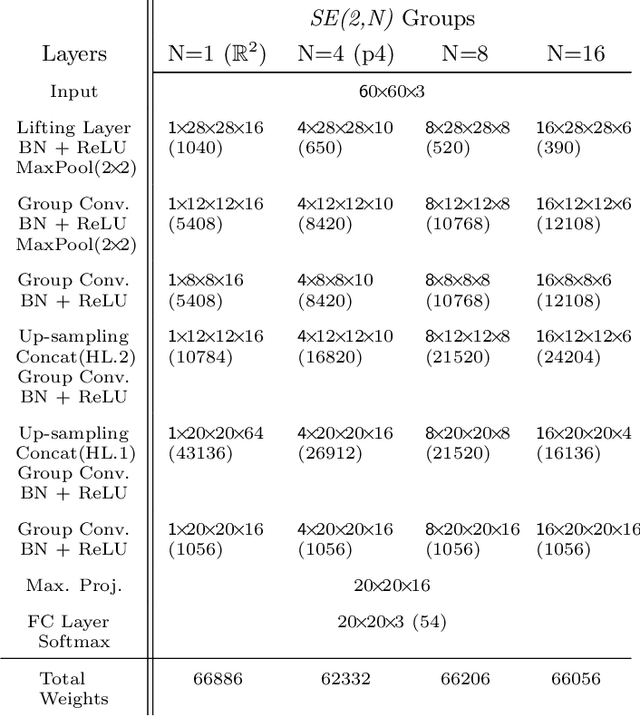
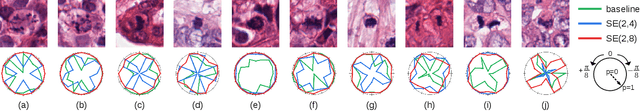
Rotation-invariance is a desired property of machine-learning models for medical image analysis and in particular for computational pathology applications. We propose a framework to encode the geometric structure of the special Euclidean motion group SE(2) in convolutional networks to yield translation and rotation equivariance via the introduction of SE(2)-group convolution layers. This structure enables models to learn feature representations with a discretized orientation dimension that guarantees that their outputs are invariant under a discrete set of rotations. Conventional approaches for rotation invariance rely mostly on data augmentation, but this does not guarantee the robustness of the output when the input is rotated. At that, trained conventional CNNs may require test-time rotation augmentation to reach their full capability. This study is focused on histopathology image analysis applications for which it is desirable that the arbitrary global orientation information of the imaged tissues is not captured by the machine learning models. The proposed framework is evaluated on three different histopathology image analysis tasks (mitosis detection, nuclei segmentation and tumor classification). We present a comparative analysis for each problem and show that consistent increase of performances can be achieved when using the proposed framework.
Direct Classification of Type 2 Diabetes From Retinal Fundus Images in a Population-based Sample From The Maastricht Study
Nov 22, 2019Friso G. Heslinga, Josien P. W. Pluim, A. J. H. M. Houben, Miranda T. Schram, Ronald M. A. Henry, Coen D. A. Stehouwer, Marleen J. van Greevenbroek, Tos T. J. M. Berendschot, Mitko Veta
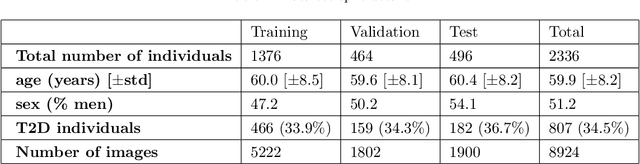
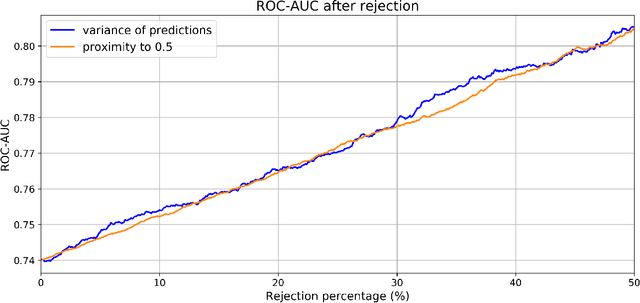
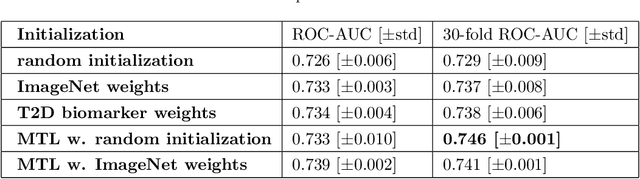

Type 2 Diabetes (T2D) is a chronic metabolic disorder that can lead to blindness and cardiovascular disease. Information about early stage T2D might be present in retinal fundus images, but to what extent these images can be used for a screening setting is still unknown. In this study, deep neural networks were employed to differentiate between fundus images from individuals with and without T2D. We investigated three methods to achieve high classification performance, measured by the area under the receiver operating curve (ROC-AUC). A multi-target learning approach to simultaneously output retinal biomarkers as well as T2D works best (AUC = 0.746 [$\pm$0.001]). Furthermore, the classification performance can be improved when images with high prediction uncertainty are referred to a specialist. We also show that the combination of images of the left and right eye per individual can further improve the classification performance (AUC = 0.758 [$\pm$0.003]), using a simple averaging approach. The results are promising, suggesting the feasibility of screening for T2D from retinal fundus images.
Deep learning assessment of breast terminal duct lobular unit involution: towards automated prediction of breast cancer risk
Oct 31, 2019Suzanne C Wetstein, Allison M Onken, Christina Luffman, Gabrielle M Baker, Michael E Pyle, Kevin H Kensler, Ying Liu, Bart Bakker, Ruud Vlutters, Marinus B van Leeuwen, Laura C Collins, Stuart J Schnitt, Josien PW Pluim, Rulla M Tamimi, Yujing J Heng, Mitko Veta
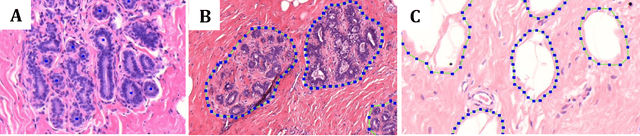
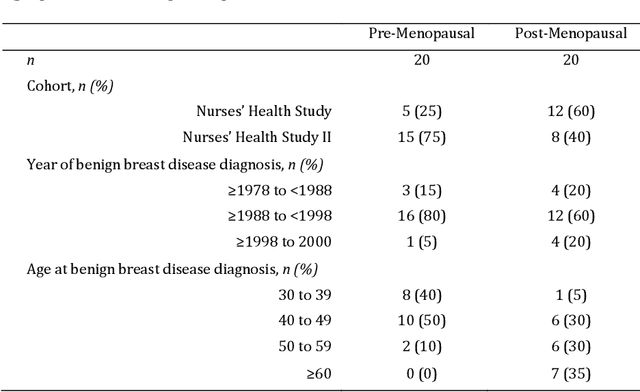
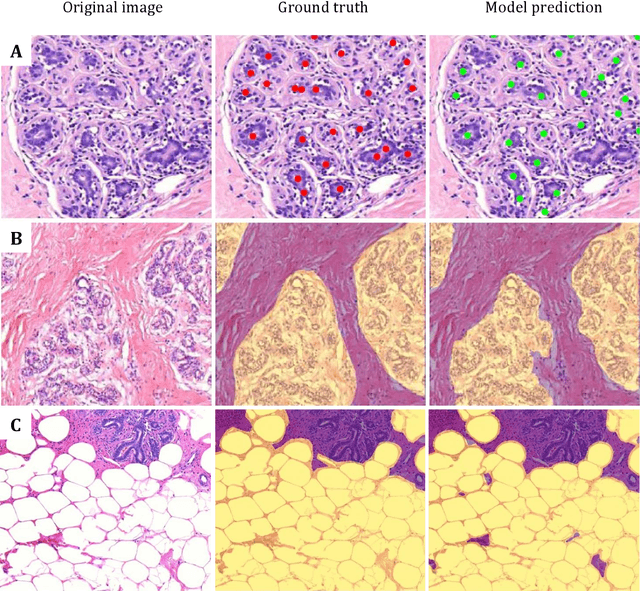
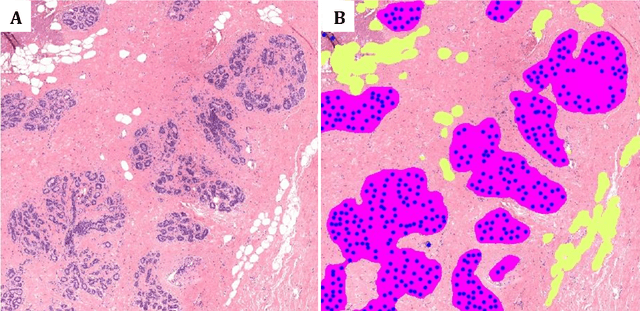
Terminal ductal lobular unit (TDLU) involution is the regression of milk-producing structures in the breast. Women with less TDLU involution are more likely to develop breast cancer. A major bottleneck in studying TDLU involution in large cohort studies is the need for labor-intensive manual assessment of TDLUs. We developed a computational pathology solution to automatically capture TDLU involution measures. Whole slide images (WSIs) of benign breast biopsies were obtained from the Nurses' Health Study (NHS). A first set of 92 WSIs was annotated for TDLUs, acini and adipose tissue to train deep convolutional neural network (CNN) models for detection of acini, and segmentation of TDLUs and adipose tissue. These networks were integrated into a single computational method to capture TDLU involution measures including number of TDLUs per tissue area, median TDLU span and median number of acini per TDLU. We validated our method on 40 additional WSIs by comparing with manually acquired measures. Our CNN models detected acini with an F1 score of 0.73$\pm$0.09, and segmented TDLUs and adipose tissue with Dice scores of 0.86$\pm$0.11 and 0.86$\pm$0.04, respectively. The inter-observer ICC scores for manual assessments on 40 WSIs of number of TDLUs per tissue area, median TDLU span, and median acini count per TDLU were 0.71, 95% CI [0.51, 0.83], 0.81, 95% CI [0.67, 0.90], and 0.73, 95% CI [0.54, 0.85], respectively. Intra-observer reliability was evaluated on 10/40 WSIs with ICC scores of >0.8. Inter-observer ICC scores between automated results and the mean of the two observers were: 0.80, 95% CI [0.63, 0.90] for number of TDLUs per tissue area, 0.57, 95% CI [0.19, 0.77] for median TDLU span, and 0.80, 95% CI [0.62, 0.89] for median acini count per TDLU. TDLU involution measures evaluated by manual and automated assessment were inversely associated with age and menopausal status.
Intensity augmentation for domain transfer of whole breast segmentation in MRI
Sep 05, 2019Linde S. Hesse, Grey Kuling, Mitko Veta, Anne L. Martel
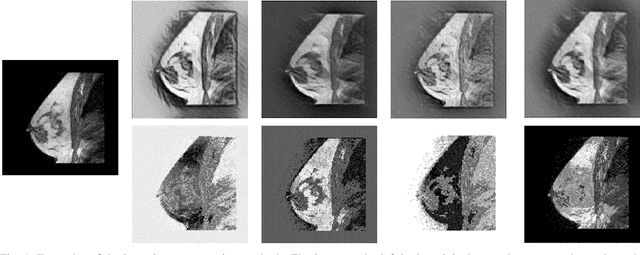
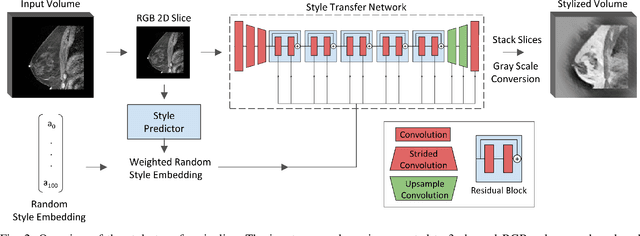
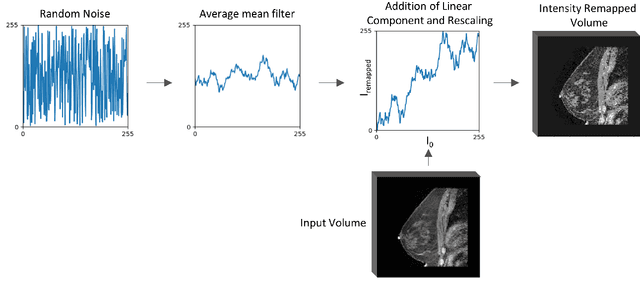
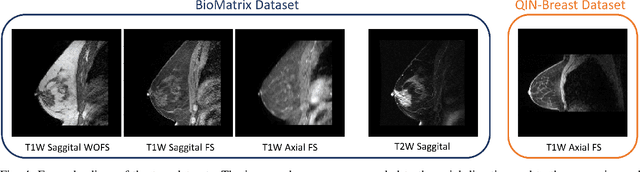
The segmentation of the breast from the chest wall is an important first step in the analysis of breast magnetic resonance images. 3D U-nets have been shown to obtain high segmentation accuracy and appear to generalize well when trained on one scanner type and tested on another scanner, provided that a very similar T1-weighted MR protocol is used. There has, however, been little work addressing the problem of domain adaptation when image intensities or patient orientation differ markedly between the training set and an unseen test set. To overcome the domain shift we propose to apply extensive intensity augmentation in addition to geometric augmentation during training. We explored both style transfer and a novel intensity remapping approach as intensity augmentation strategies. For our experiments, we trained a 3D U-net on T1-weighted scans and tested on T2-weighted scans. By applying intensity augmentation we increased segmentation performance from a DSC of 0.71 to 0.90. This performance is very close to the baseline performance of training and testing on T2-weighted scans (0.92). Furthermore, we applied our network to an independent test set made up of publicly available scans acquired using a T1-weighted TWIST sequence and a different coil configuration. On this dataset we obtained a performance of 0.89, close to the inter-observer variability of the ground truth segmentations (0.92). Our results show that using intensity augmentation in addition to geometric augmentation is a suitable method to overcome the intensity domain shift and we expect it to be useful for a wide range of segmentation tasks.
Deep learning-based prediction of kinetic parameters from myocardial perfusion MRI
Jul 27, 2019Cian M. Scannell, Piet van den Bosch, Amedeo Chiribiri, Jack Lee, Marcel Breeuwer, Mitko Veta
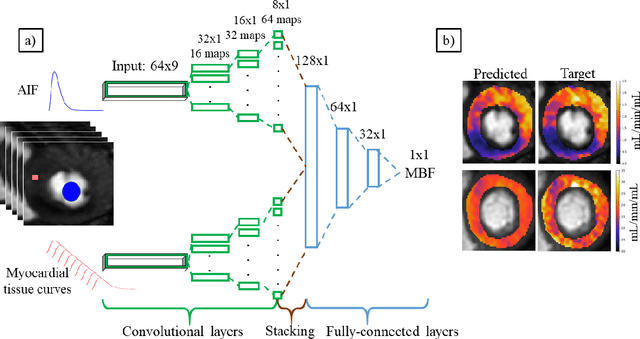
The quantification of myocardial perfusion MRI has the potential to provide a fast, automated and user-independent assessment of myocardial ischaemia. However, due to the relatively high noise level and low temporal resolution of the acquired data and the complexity of the tracer-kinetic models, the model fitting can yield unreliable parameter estimates. A solution to this problem is the use of Bayesian inference which can incorporate prior knowledge and improve the reliability of the parameter estimation. This, however, uses Markov chain Monte Carlo sampling to approximate the posterior distribution of the kinetic parameters which is extremely time intensive. This work proposes training convolutional networks to directly predict the kinetic parameters from the signal-intensity curves that are trained using estimates obtained from the Bayesian inference. This allows fast estimation of the kinetic parameters with a similar performance to the Bayesian inference.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge
Jul 22, 2018Mitko Veta, Yujing J. Heng, Nikolas Stathonikos, Babak Ehteshami Bejnordi, Francisco Beca, Thomas Wollmann, Karl Rohr, Manan A. Shah, Dayong Wang, Mikael Rousson, Martin Hedlund, David Tellez, Francesco Ciompi, Erwan Zerhouni, David Lanyi, Matheus Viana, Vassili Kovalev, Vitali Liauchuk, Hady Ahmady Phoulady, Talha Qaiser, Simon Graham, Nasir Rajpoot, Erik Sjöblom, Jesper Molin, Kyunghyun Paeng, Sangheum Hwang, Sunggyun Park, Zhipeng Jia, Eric I-Chao Chang, Yan Xu, Andrew H. Beck, Paul J. van Diest, Josien P. W. Pluim
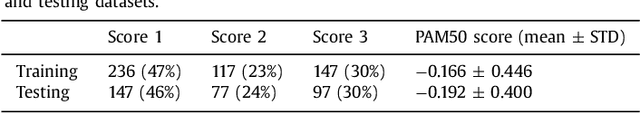
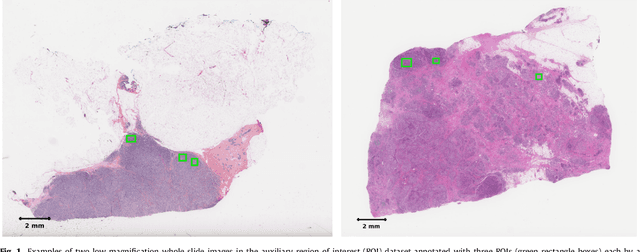
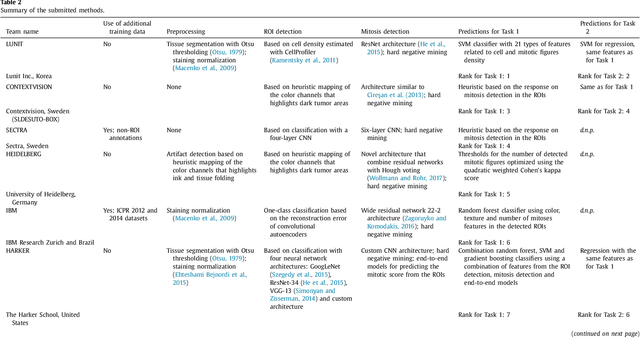
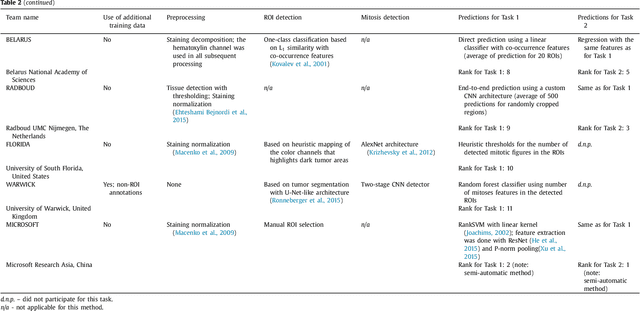
Tumor proliferation is an important biomarker indicative of the prognosis of breast cancer patients. Assessment of tumor proliferation in a clinical setting is highly subjective and labor-intensive task. Previous efforts to automate tumor proliferation assessment by image analysis only focused on mitosis detection in predefined tumor regions. However, in a real-world scenario, automatic mitosis detection should be performed in whole-slide images (WSIs) and an automatic method should be able to produce a tumor proliferation score given a WSI as input. To address this, we organized the TUmor Proliferation Assessment Challenge 2016 (TUPAC16) on prediction of tumor proliferation scores from WSIs. The challenge dataset consisted of 500 training and 321 testing breast cancer histopathology WSIs. In order to ensure fair and independent evaluation, only the ground truth for the training dataset was provided to the challenge participants. The first task of the challenge was to predict mitotic scores, i.e., to reproduce the manual method of assessing tumor proliferation by a pathologist. The second task was to predict the gene expression based PAM50 proliferation scores from the WSI. The best performing automatic method for the first task achieved a quadratic-weighted Cohen's kappa score of $\kappa$ = 0.567, 95% CI [0.464, 0.671] between the predicted scores and the ground truth. For the second task, the predictions of the top method had a Spearman's correlation coefficient of r = 0.617, 95% CI [0.581 0.651] with the ground truth. This was the first study that investigated tumor proliferation assessment from WSIs. The achieved results are promising given the difficulty of the tasks and weakly-labelled nature of the ground truth. However, further research is needed to improve the practical utility of image analysis methods for this task.
Roto-Translation Covariant Convolutional Networks for Medical Image Analysis
Jun 11, 2018Erik J Bekkers, Maxime W Lafarge, Mitko Veta, Koen AJ Eppenhof, Josien PW Pluim, Remco Duits
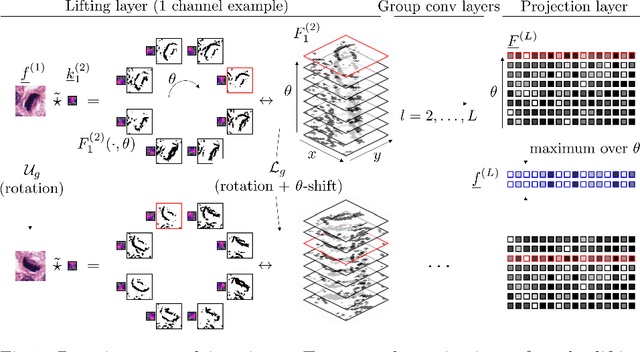
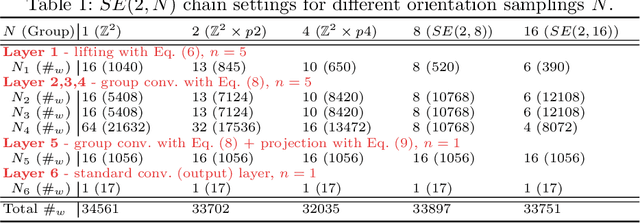
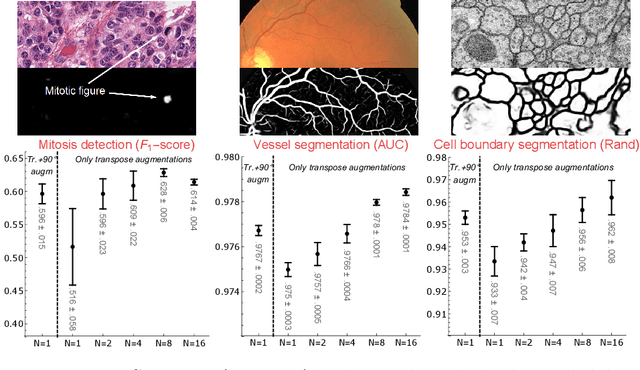
We propose a framework for rotation and translation covariant deep learning using $SE(2)$ group convolutions. The group product of the special Euclidean motion group $SE(2)$ describes how a concatenation of two roto-translations results in a net roto-translation. We encode this geometric structure into convolutional neural networks (CNNs) via $SE(2)$ group convolutional layers, which fit into the standard 2D CNN framework, and which allow to generically deal with rotated input samples without the need for data augmentation. We introduce three layers: a lifting layer which lifts a 2D (vector valued) image to an $SE(2)$-image, i.e., 3D (vector valued) data whose domain is $SE(2)$; a group convolution layer from and to an $SE(2)$-image; and a projection layer from an $SE(2)$-image to a 2D image. The lifting and group convolution layers are $SE(2)$ covariant (the output roto-translates with the input). The final projection layer, a maximum intensity projection over rotations, makes the full CNN rotation invariant. We show with three different problems in histopathology, retinal imaging, and electron microscopy that with the proposed group CNNs, state-of-the-art performance can be achieved, without the need for data augmentation by rotation and with increased performance compared to standard CNNs that do rely on augmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge