Eric I-Chao Chang
Exploring Diffusion Time-steps for Unsupervised Representation Learning
Jan 21, 2024Abstract:Representation learning is all about discovering the hidden modular attributes that generate the data faithfully. We explore the potential of Denoising Diffusion Probabilistic Model (DM) in unsupervised learning of the modular attributes. We build a theoretical framework that connects the diffusion time-steps and the hidden attributes, which serves as an effective inductive bias for unsupervised learning. Specifically, the forward diffusion process incrementally adds Gaussian noise to samples at each time-step, which essentially collapses different samples into similar ones by losing attributes, e.g., fine-grained attributes such as texture are lost with less noise added (i.e., early time-steps), while coarse-grained ones such as shape are lost by adding more noise (i.e., late time-steps). To disentangle the modular attributes, at each time-step t, we learn a t-specific feature to compensate for the newly lost attribute, and the set of all 1,...,t-specific features, corresponding to the cumulative set of lost attributes, are trained to make up for the reconstruction error of a pre-trained DM at time-step t. On CelebA, FFHQ, and Bedroom datasets, the learned feature significantly improves attribute classification and enables faithful counterfactual generation, e.g., interpolating only one specified attribute between two images, validating the disentanglement quality. Codes are in https://github.com/yue-zhongqi/diti.
EHRXQA: A Multi-Modal Question Answering Dataset for Electronic Health Records with Chest X-ray Images
Oct 28, 2023



Abstract:Electronic Health Records (EHRs), which contain patients' medical histories in various multi-modal formats, often overlook the potential for joint reasoning across imaging and table modalities underexplored in current EHR Question Answering (QA) systems. In this paper, we introduce EHRXQA, a novel multi-modal question answering dataset combining structured EHRs and chest X-ray images. To develop our dataset, we first construct two uni-modal resources: 1) The MIMIC- CXR-VQA dataset, our newly created medical visual question answering (VQA) benchmark, specifically designed to augment the imaging modality in EHR QA, and 2) EHRSQL (MIMIC-IV), a refashioned version of a previously established table-based EHR QA dataset. By integrating these two uni-modal resources, we successfully construct a multi-modal EHR QA dataset that necessitates both uni-modal and cross-modal reasoning. To address the unique challenges of multi-modal questions within EHRs, we propose a NeuralSQL-based strategy equipped with an external VQA API. This pioneering endeavor enhances engagement with multi-modal EHR sources and we believe that our dataset can catalyze advances in real-world medical scenarios such as clinical decision-making and research. EHRXQA is available at https://github.com/baeseongsu/ehrxqa.
3D Segmentation Guided Style-based Generative Adversarial Networks for PET Synthesis
May 18, 2022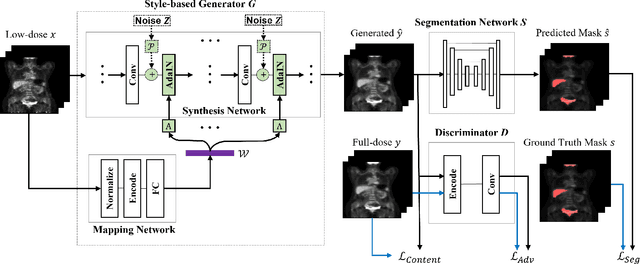
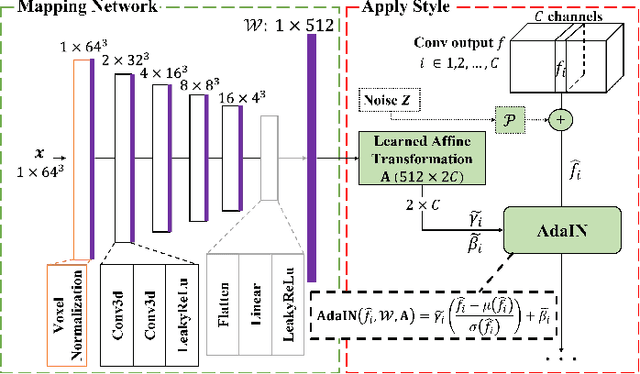
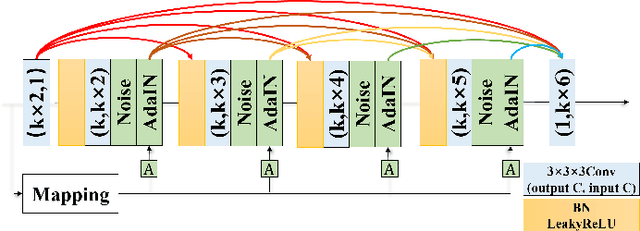
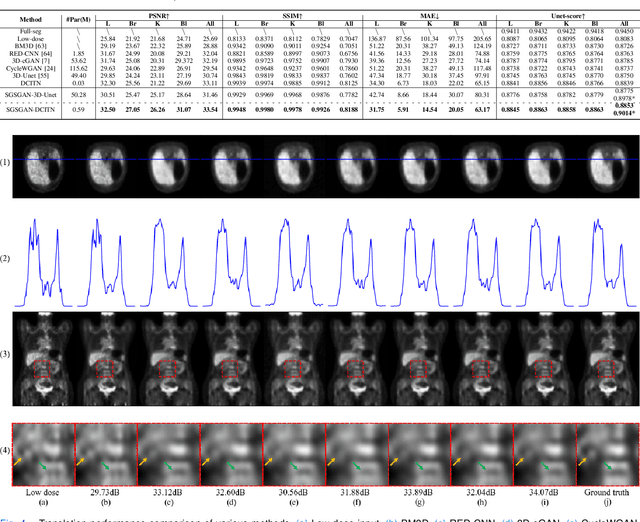
Abstract:Potential radioactive hazards in full-dose positron emission tomography (PET) imaging remain a concern, whereas the quality of low-dose images is never desirable for clinical use. So it is of great interest to translate low-dose PET images into full-dose. Previous studies based on deep learning methods usually directly extract hierarchical features for reconstruction. We notice that the importance of each feature is different and they should be weighted dissimilarly so that tiny information can be captured by the neural network. Furthermore, the synthesis on some regions of interest is important in some applications. Here we propose a novel segmentation guided style-based generative adversarial network (SGSGAN) for PET synthesis. (1) We put forward a style-based generator employing style modulation, which specifically controls the hierarchical features in the translation process, to generate images with more realistic textures. (2) We adopt a task-driven strategy that couples a segmentation task with a generative adversarial network (GAN) framework to improve the translation performance. Extensive experiments show the superiority of our overall framework in PET synthesis, especially on those regions of interest.
Transformer based multiple instance learning for weakly supervised histopathology image segmentation
May 18, 2022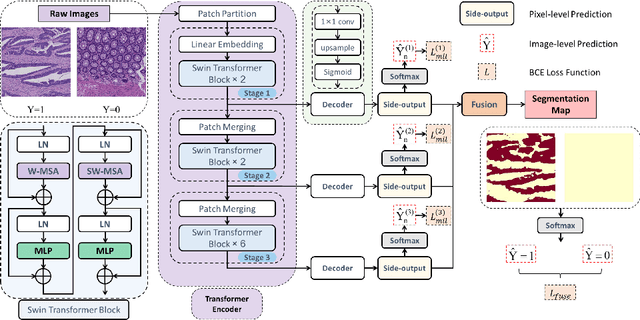
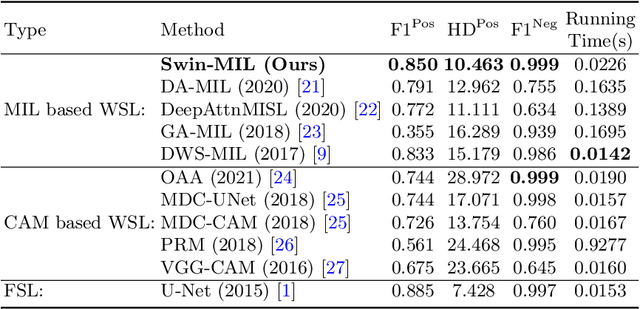
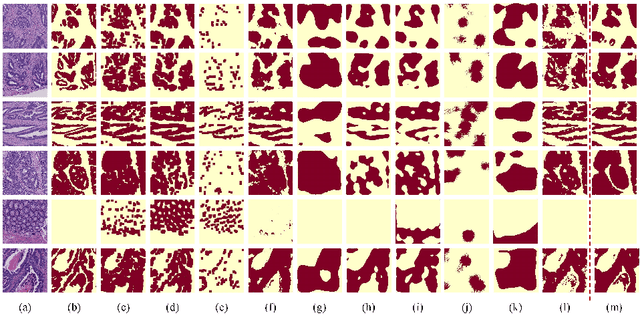
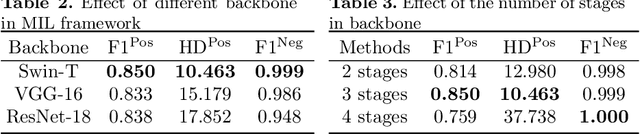
Abstract:Hispathological image segmentation algorithms play a critical role in computer aided diagnosis technology. The development of weakly supervised segmentation algorithm alleviates the problem of medical image annotation that it is time-consuming and labor-intensive. As a subset of weakly supervised learning, Multiple Instance Learning (MIL) has been proven to be effective in segmentation. However, there is a lack of related information between instances in MIL, which limits the further improvement of segmentation performance. In this paper, we propose a novel weakly supervised method for pixel-level segmentation in histopathology images, which introduces Transformer into the MIL framework to capture global or long-range dependencies. The multi-head self-attention in the Transformer establishes the relationship between instances, which solves the shortcoming that instances are independent of each other in MIL. In addition, deep supervision is introduced to overcome the limitation of annotations in weakly supervised methods and make the better utilization of hierarchical information. The state-of-the-art results on the colon cancer dataset demonstrate the superiority of the proposed method compared with other weakly supervised methods. It is worth believing that there is a potential of our approach for various applications in medical images.
Whole Brain Segmentation with Full Volume Neural Network
Oct 29, 2021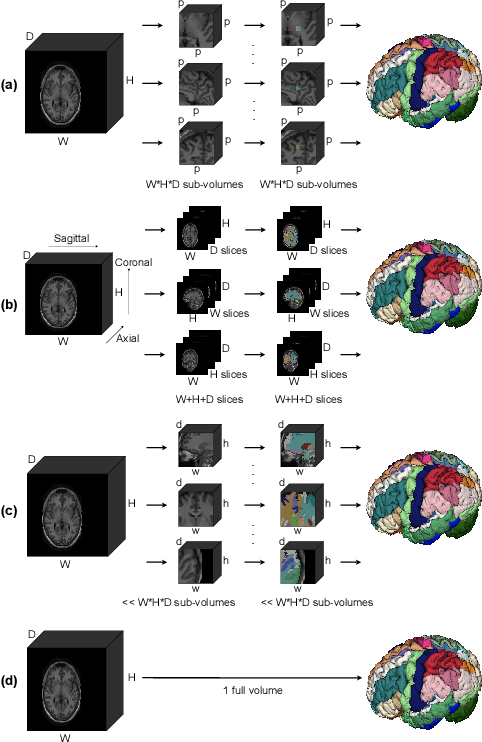
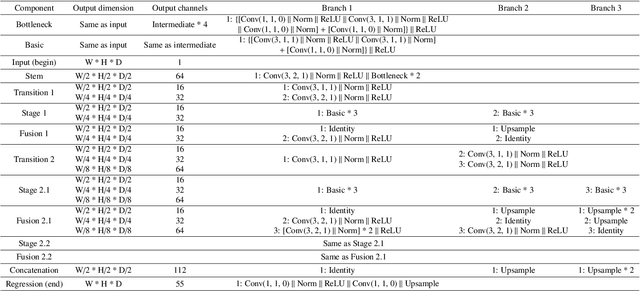
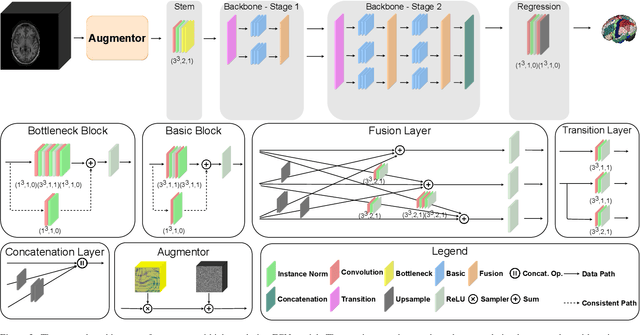
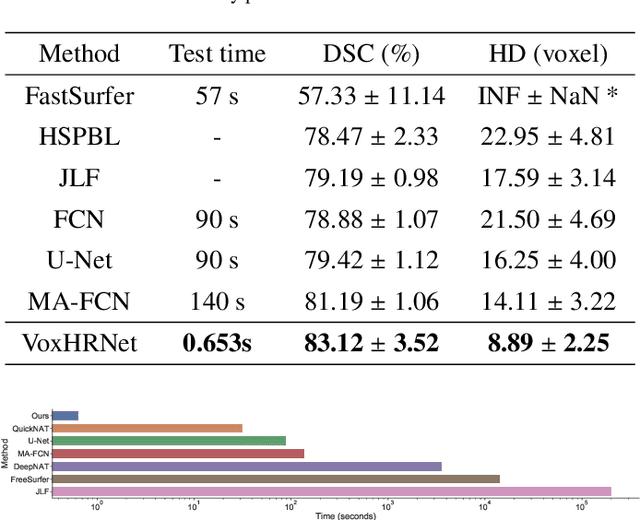
Abstract:Whole brain segmentation is an important neuroimaging task that segments the whole brain volume into anatomically labeled regions-of-interest. Convolutional neural networks have demonstrated good performance in this task. Existing solutions, usually segment the brain image by classifying the voxels, or labeling the slices or the sub-volumes separately. Their representation learning is based on parts of the whole volume whereas their labeling result is produced by aggregation of partial segmentation. Learning and inference with incomplete information could lead to sub-optimal final segmentation result. To address these issues, we propose to adopt a full volume framework, which feeds the full volume brain image into the segmentation network and directly outputs the segmentation result for the whole brain volume. The framework makes use of complete information in each volume and can be implemented easily. An effective instance in this framework is given subsequently. We adopt the $3$D high-resolution network (HRNet) for learning spatially fine-grained representations and the mixed precision training scheme for memory-efficient training. Extensive experiment results on a publicly available $3$D MRI brain dataset show that our proposed model advances the state-of-the-art methods in terms of segmentation performance. Source code is publicly available at https://github.com/microsoft/VoxHRNet.
* Accepted to CMIG
Microscopic fine-grained instance classification through deep attention
Oct 06, 2020
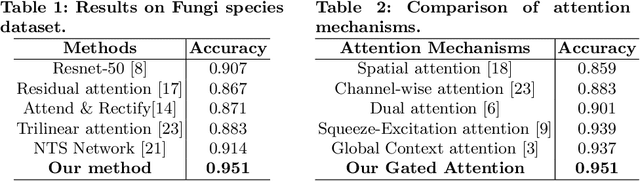
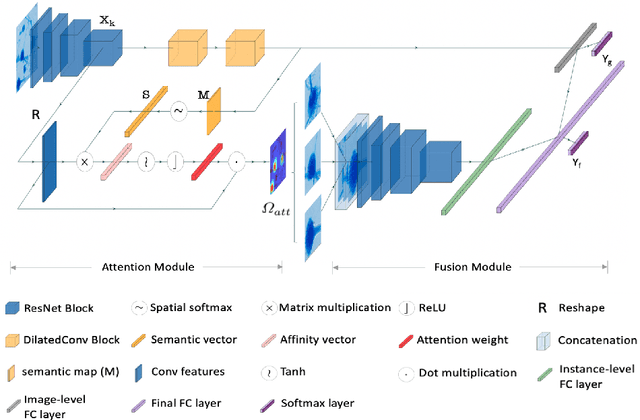
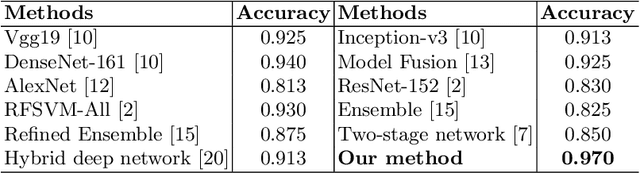
Abstract:Fine-grained classification of microscopic image data with limited samples is an open problem in computer vision and biomedical imaging. Deep learning based vision systems mostly deal with high number of low-resolution images, whereas subtle detail in biomedical images require higher resolution. To bridge this gap, we propose a simple yet effective deep network that performs two tasks simultaneously in an end-to-end manner. First, it utilises a gated attention module that can focus on multiple key instances at high resolution without extra annotations or region proposals. Second, the global structural features and local instance features are fused for final image level classification. The result is a robust but lightweight end-to-end trainable deep network that yields state-of-the-art results in two separate fine-grained multi-instance biomedical image classification tasks: a benchmark breast cancer histology dataset and our new fungi species mycology dataset. In addition, we demonstrate the interpretability of the proposed model by visualising the concordance of the learned features with clinically relevant features.
MaskFlownet: Asymmetric Feature Matching with Learnable Occlusion Mask
Apr 08, 2020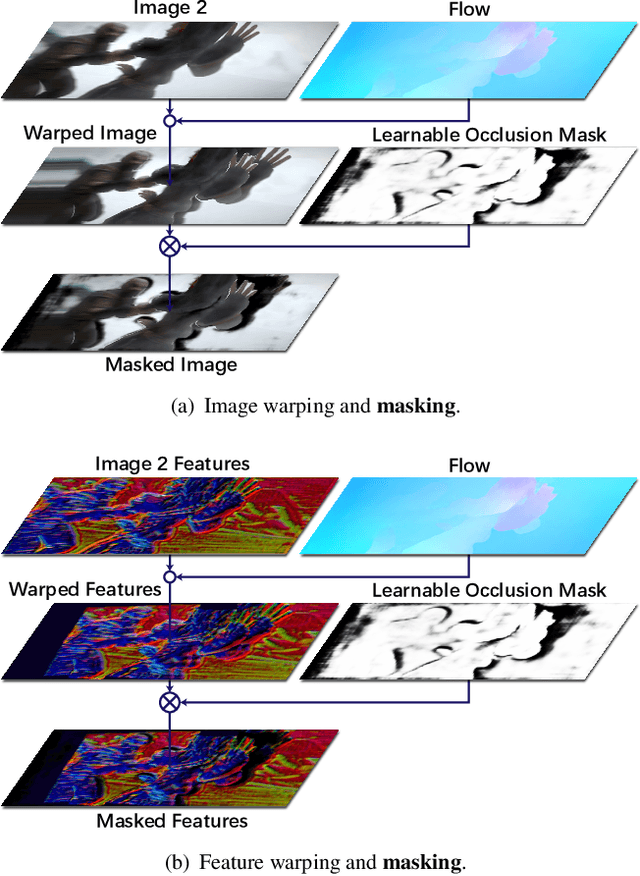
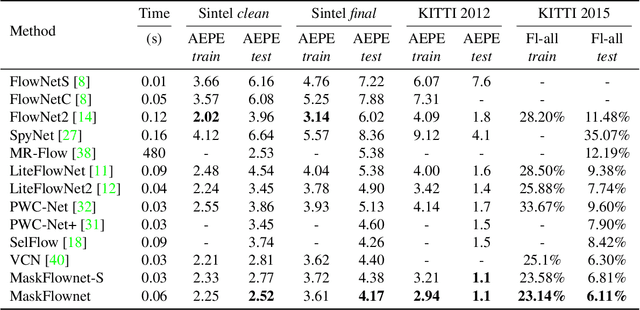
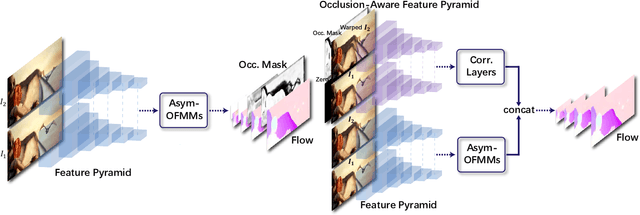
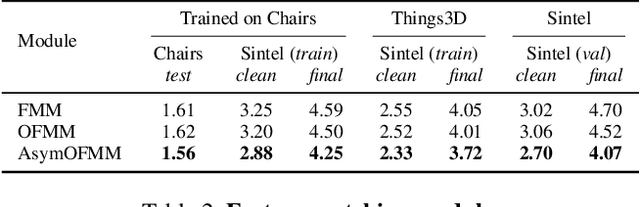
Abstract:Feature warping is a core technique in optical flow estimation; however, the ambiguity caused by occluded areas during warping is a major problem that remains unsolved. In this paper, we propose an asymmetric occlusion-aware feature matching module, which can learn a rough occlusion mask that filters useless (occluded) areas immediately after feature warping without any explicit supervision. The proposed module can be easily integrated into end-to-end network architectures and enjoys performance gains while introducing negligible computational cost. The learned occlusion mask can be further fed into a subsequent network cascade with dual feature pyramids with which we achieve state-of-the-art performance. At the time of submission, our method, called MaskFlownet, surpasses all published optical flow methods on the MPI Sintel, KITTI 2012 and 2015 benchmarks. Code is available at https://github.com/microsoft/MaskFlownet.
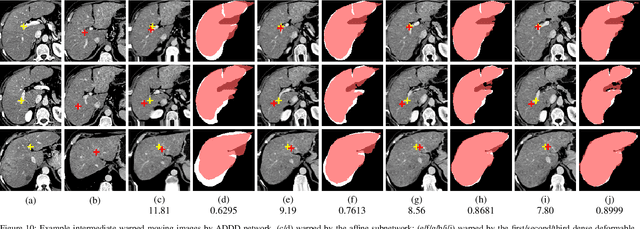
Recursive Cascaded Networks for Unsupervised Medical Image Registration
Jul 29, 2019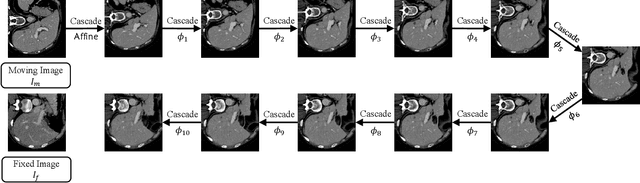

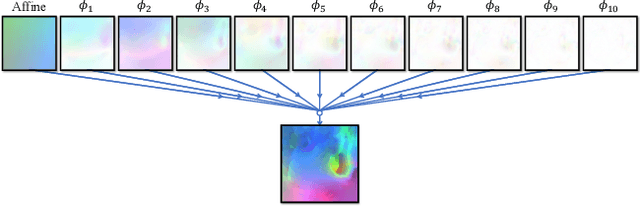
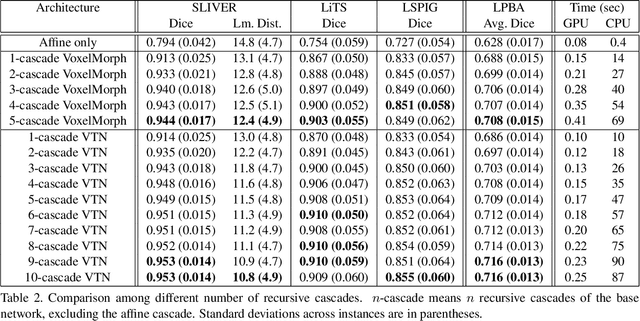
Abstract:We present recursive cascaded networks, a general architecture that enables learning deep cascades, for deformable image registration. The proposed architecture is simple in design and can be built on any base network. The moving image is warped successively by each cascade and finally aligned to the fixed image; this procedure is recursive in a way that every cascade learns to perform a progressive deformation for the current warped image. The entire system is end-to-end and jointly trained in an unsupervised manner. In addition, enabled by the recursive architecture, one cascade can be iteratively applied for multiple times during testing, which approaches a better fit between each of the image pairs. We evaluate our method on 3D medical images, where deformable registration is most commonly applied. We demonstrate that recursive cascaded networks achieve consistent, significant gains and outperform state-of-the-art methods. The performance reveals an increasing trend as long as more cascades are trained, while the limit is not observed. Our code will be made publicly available.
Unsupervised 3D End-to-End Medical Image Registration with Volume Tweening Network
Feb 13, 2019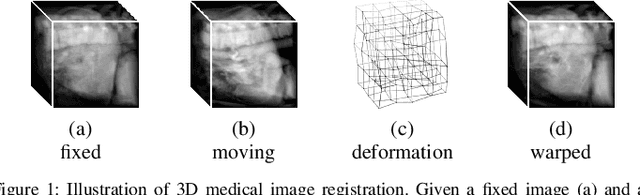
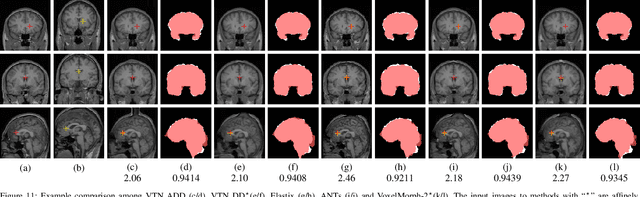
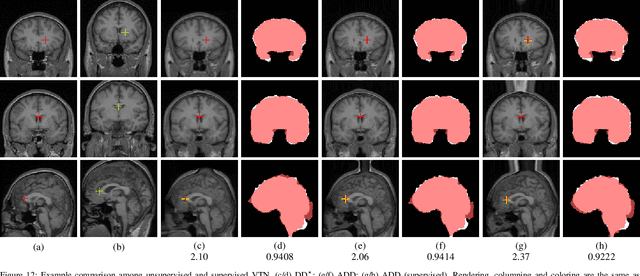
Abstract:3D medical image registration is of great clinical importance. However, supervised learning methods require a large amount of accurately annotated corresponding control points (or morphing). The ground truth for 3D medical images is very difficult to obtain. Unsupervised learning methods ease the burden of manual annotation by exploiting unlabeled data without supervision. In this paper, we propose a new unsupervised learning method using convolutional neural networks under an end-to-end framework, Volume Tweening Network (VTN), to register 3D medical images. Three technical components ameliorate our unsupervised learning system for 3D end-to-end medical image registration: (1) We cascade the registration subnetworks; (2) We integrate affine registration into our network; and (3) We incorporate an additional invertibility loss into the training process. Experimental results demonstrate that our algorithm is 880x faster (or 3.3x faster without GPU acceleration) than traditional optimization-based methods and achieves state-of-the-art performance in medical image registration.
MRI Cross-Modality NeuroImage-to-NeuroImage Translation
Sep 11, 2018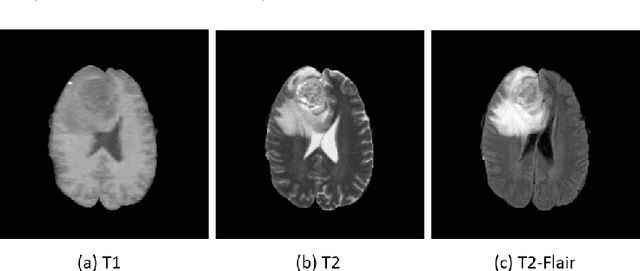
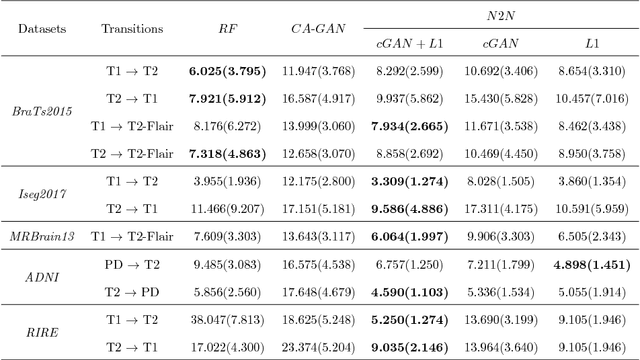

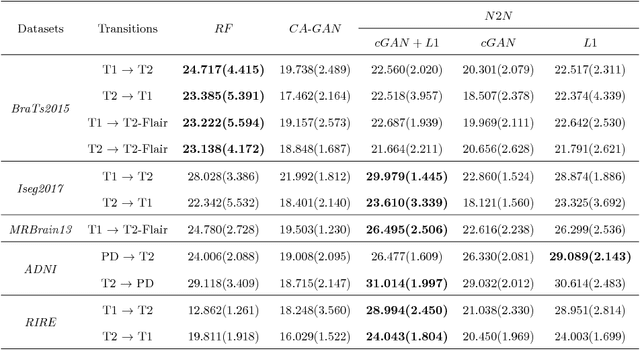
Abstract:We present a cross-modality generation framework that learns to generate translated modalities from given modalities in MR images without real acquisition. Our proposed method performs NeuroImage-to-NeuroImage translation (abbreviated as N2N) by means of a deep learning model that leverages conditional generative adversarial networks (cGANs). Our framework jointly exploits the low-level features (pixel-wise information) and high-level representations (e.g. brain tumors, brain structure like gray matter, etc.) between cross modalities which are important for resolving the challenging complexity in brain structures. Our framework can serve as an auxiliary method in clinical diagnosis and has great application potential. Based on our proposed framework, we first propose a method for cross-modality registration by fusing the deformation fields to adopt the cross-modality information from translated modalities. Second, we propose an approach for MRI segmentation, translated multichannel segmentation (TMS), where given modalities, along with translated modalities, are segmented by fully convolutional networks (FCN) in a multichannel manner. Both of these two methods successfully adopt the cross-modality information to improve the performance without adding any extra data. Experiments demonstrate that our proposed framework advances the state-of-the-art on five brain MRI datasets. We also observe encouraging results in cross-modality registration and segmentation on some widely adopted brain datasets. Overall, our work can serve as an auxiliary method in clinical diagnosis and be applied to various tasks in medical fields. Keywords: image-to-image, cross-modality, registration, segmentation, brain MRI
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge