Zhipeng Jia
Spark Transformer: Reactivating Sparsity in FFN and Attention
Jun 07, 2025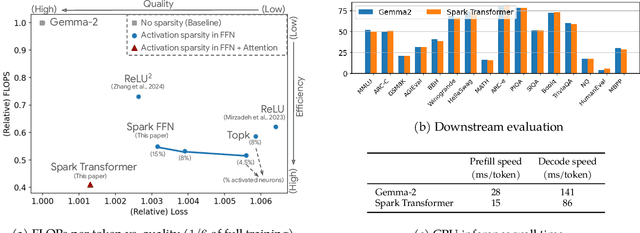
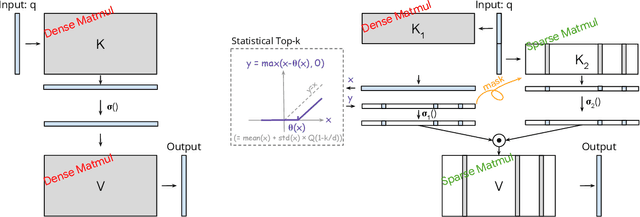
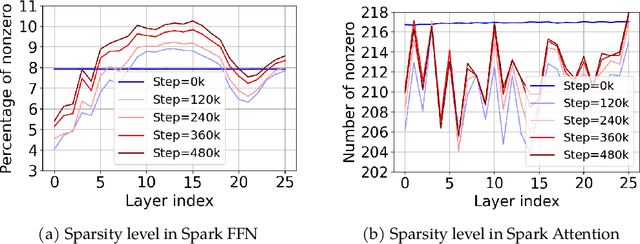
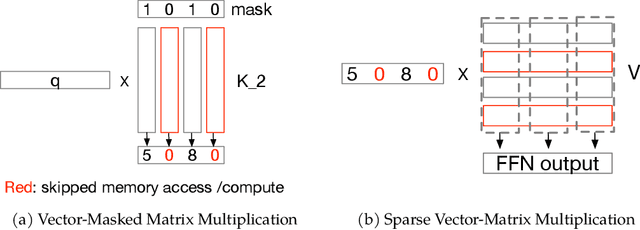
Abstract:The discovery of the lazy neuron phenomenon in trained Transformers, where the vast majority of neurons in their feed-forward networks (FFN) are inactive for each token, has spurred tremendous interests in activation sparsity for enhancing large model efficiency. While notable progress has been made in translating such sparsity to wall-time benefits, modern Transformers have moved away from the ReLU activation function crucial to this phenomenon. Existing efforts on re-introducing activation sparsity often degrade model quality, increase parameter count, complicate or slow down training. Sparse attention, the application of sparse activation to the attention mechanism, often faces similar challenges. This paper introduces the Spark Transformer, a novel architecture that achieves a high level of activation sparsity in both FFN and the attention mechanism while maintaining model quality, parameter count, and standard training procedures. Our method realizes sparsity via top-k masking for explicit control over sparsity level. Crucially, we introduce statistical top-k, a hardware-accelerator-friendly, linear-time approximate algorithm that avoids costly sorting and mitigates significant training slowdown from standard top-$k$ operators. Furthermore, Spark Transformer reallocates existing FFN parameters and attention key embeddings to form a low-cost predictor for identifying activated entries. This design not only mitigates quality loss from enforced sparsity, but also enhances wall-time benefit. Pretrained with the Gemma-2 recipe, Spark Transformer demonstrates competitive performance on standard benchmarks while exhibiting significant sparsity: only 8% of FFN neurons are activated, and each token attends to a maximum of 256 tokens. This sparsity translates to a 2.5x reduction in FLOPs, leading to decoding wall-time speedups of up to 1.79x on CPU and 1.40x on GPU.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge
Jul 22, 2018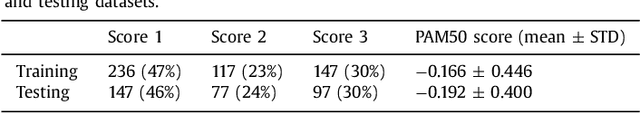
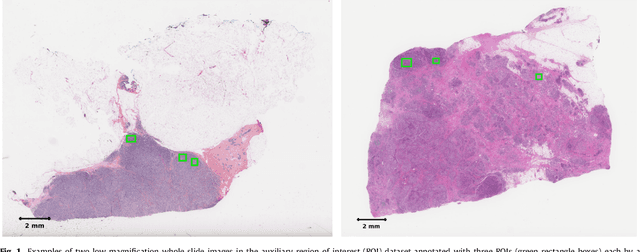
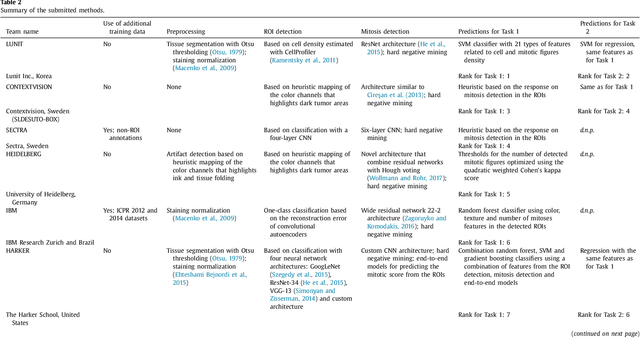
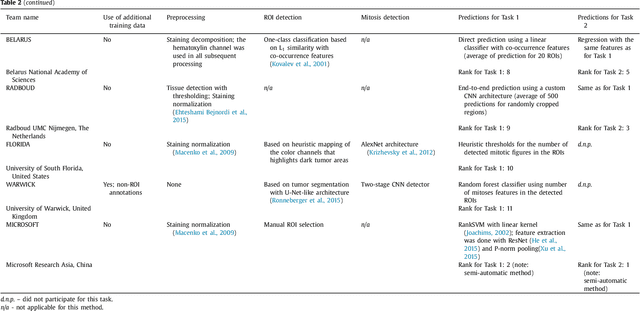
Abstract:Tumor proliferation is an important biomarker indicative of the prognosis of breast cancer patients. Assessment of tumor proliferation in a clinical setting is highly subjective and labor-intensive task. Previous efforts to automate tumor proliferation assessment by image analysis only focused on mitosis detection in predefined tumor regions. However, in a real-world scenario, automatic mitosis detection should be performed in whole-slide images (WSIs) and an automatic method should be able to produce a tumor proliferation score given a WSI as input. To address this, we organized the TUmor Proliferation Assessment Challenge 2016 (TUPAC16) on prediction of tumor proliferation scores from WSIs. The challenge dataset consisted of 500 training and 321 testing breast cancer histopathology WSIs. In order to ensure fair and independent evaluation, only the ground truth for the training dataset was provided to the challenge participants. The first task of the challenge was to predict mitotic scores, i.e., to reproduce the manual method of assessing tumor proliferation by a pathologist. The second task was to predict the gene expression based PAM50 proliferation scores from the WSI. The best performing automatic method for the first task achieved a quadratic-weighted Cohen's kappa score of $\kappa$ = 0.567, 95% CI [0.464, 0.671] between the predicted scores and the ground truth. For the second task, the predictions of the top method had a Spearman's correlation coefficient of r = 0.617, 95% CI [0.581 0.651] with the ground truth. This was the first study that investigated tumor proliferation assessment from WSIs. The achieved results are promising given the difficulty of the tasks and weakly-labelled nature of the ground truth. However, further research is needed to improve the practical utility of image analysis methods for this task.
Constrained Deep Weak Supervision for Histopathology Image Segmentation
Jan 03, 2017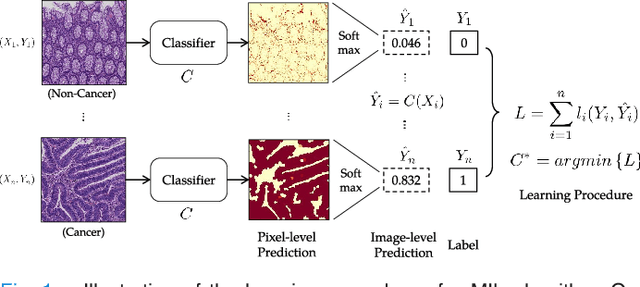
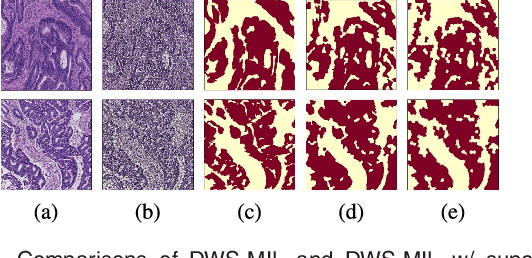
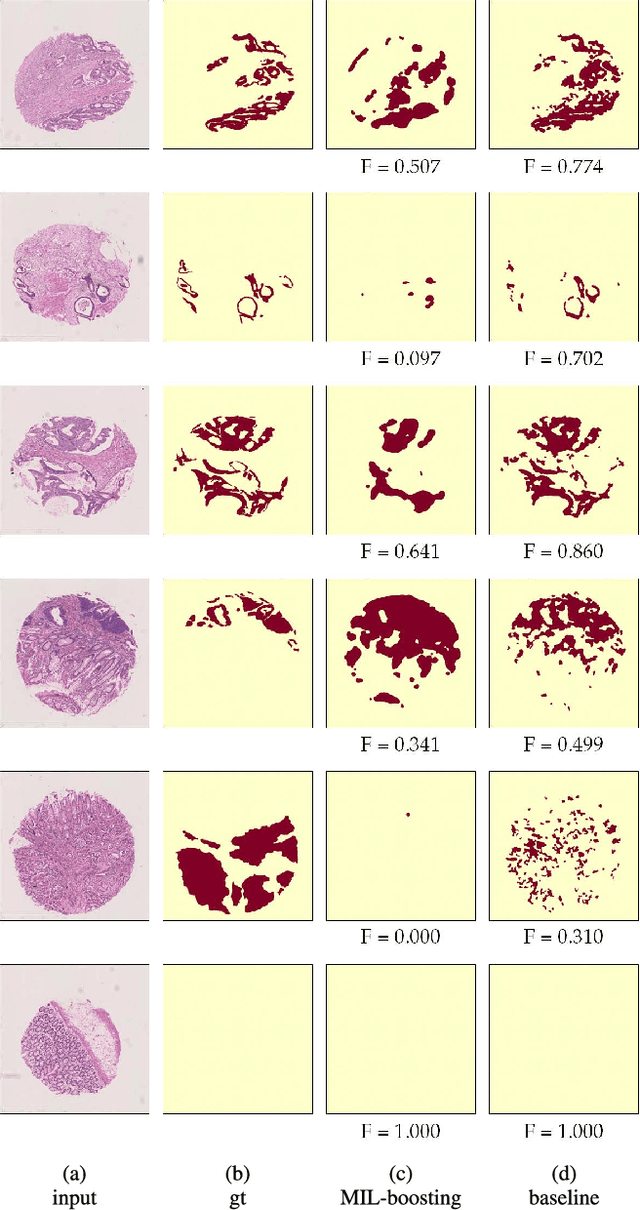
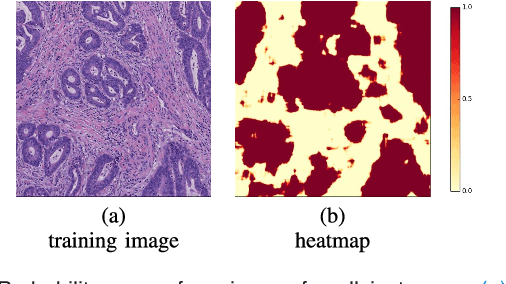
Abstract:In this paper, we develop a new weakly-supervised learning algorithm to learn to segment cancerous regions in histopathology images. Our work is under a multiple instance learning framework (MIL) with a new formulation, deep weak supervision (DWS); we also propose an effective way to introduce constraints to our neural networks to assist the learning process. The contributions of our algorithm are threefold: (1) We build an end-to-end learning system that segments cancerous regions with fully convolutional networks (FCN) in which image-to-image weakly-supervised learning is performed. (2) We develop a deep week supervision formulation to exploit multi-scale learning under weak supervision within fully convolutional networks. (3) Constraints about positive instances are introduced in our approach to effectively explore additional weakly-supervised information that is easy to obtain and enjoys a significant boost to the learning process. The proposed algorithm, abbreviated as DWS-MIL, is easy to implement and can be trained efficiently. Our system demonstrates state-of-the-art results on large-scale histopathology image datasets and can be applied to various applications in medical imaging beyond histopathology images such as MRI, CT, and ultrasound images.
End-to-End Subtitle Detection and Recognition for Videos in East Asian Languages via CNN Ensemble with Near-Human-Level Performance
Nov 18, 2016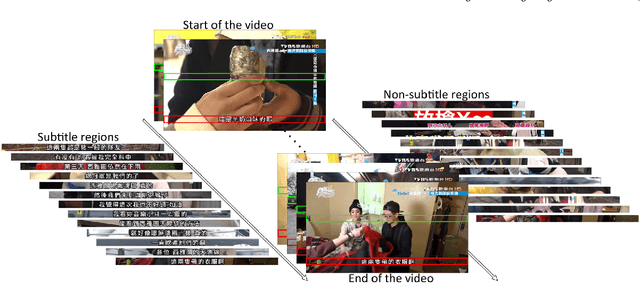
Abstract:In this paper, we propose an innovative end-to-end subtitle detection and recognition system for videos in East Asian languages. Our end-to-end system consists of multiple stages. Subtitles are firstly detected by a novel image operator based on the sequence information of consecutive video frames. Then, an ensemble of Convolutional Neural Networks (CNNs) trained on synthetic data is adopted for detecting and recognizing East Asian characters. Finally, a dynamic programming approach leveraging language models is applied to constitute results of the entire body of text lines. The proposed system achieves average end-to-end accuracies of 98.2% and 98.3% on 40 videos in Simplified Chinese and 40 videos in Traditional Chinese respectively, which is a significant outperformance of other existing methods. The near-perfect accuracy of our system dramatically narrows the gap between human cognitive ability and state-of-the-art algorithms used for such a task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge