"cancer detection": models, code, and papers
ProstAttention-Net: A deep attention model for prostate cancer segmentation by aggressiveness in MRI scans
Nov 23, 2022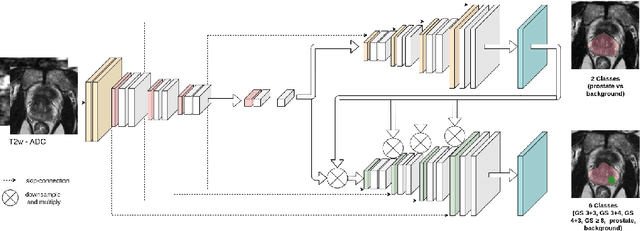
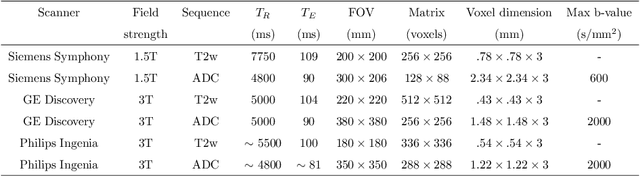
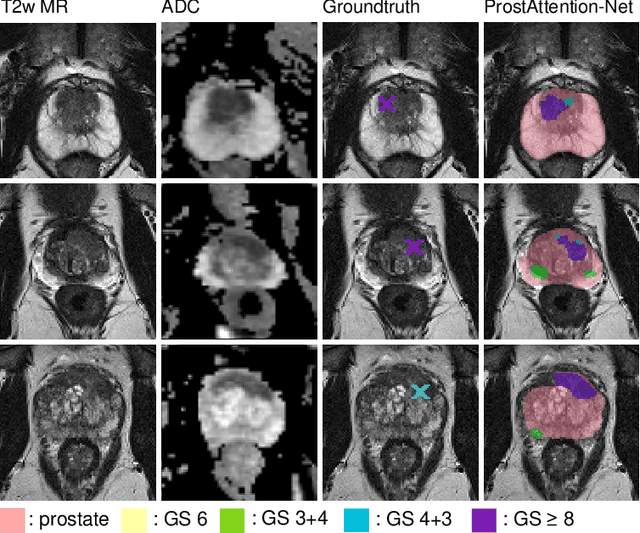
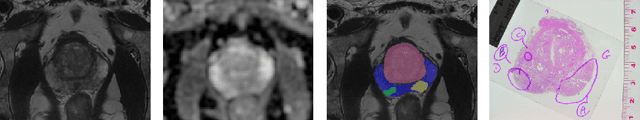
Multiparametric magnetic resonance imaging (mp-MRI) has shown excellent results in the detection of prostate cancer (PCa). However, characterizing prostate lesions aggressiveness in mp-MRI sequences is impossible in clinical practice, and biopsy remains the reference to determine the Gleason score (GS). In this work, we propose a novel end-to-end multi-class network that jointly segments the prostate gland and cancer lesions with GS group grading. After encoding the information on a latent space, the network is separated in two branches: 1) the first branch performs prostate segmentation 2) the second branch uses this zonal prior as an attention gate for the detection and grading of prostate lesions. The model was trained and validated with a 5-fold cross-validation on an heterogeneous series of 219 MRI exams acquired on three different scanners prior prostatectomy. In the free-response receiver operating characteristics (FROC) analysis for clinically significant lesions (defined as GS > 6) detection, our model achieves 69.0% $\pm$14.5% sensitivity at 2.9 false positive per patient on the whole prostate and 70.8% $\pm$14.4% sensitivity at 1.5 false positive when considering the peripheral zone (PZ) only. Regarding the automatic GS group
A Comparative Study of Gastric Histopathology Sub-size Image Classification: from Linear Regression to Visual Transformer
May 25, 2022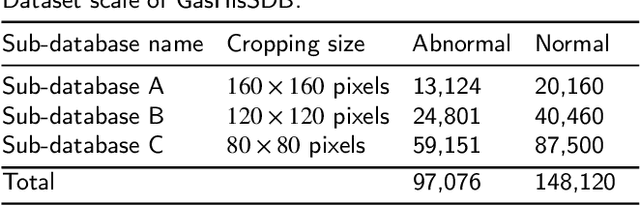
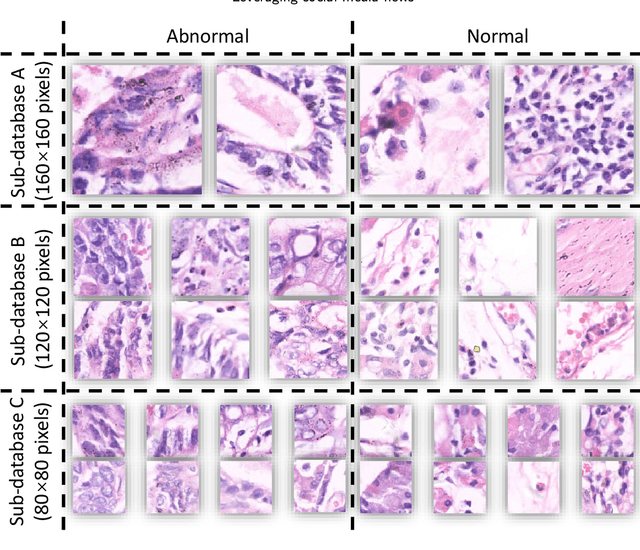
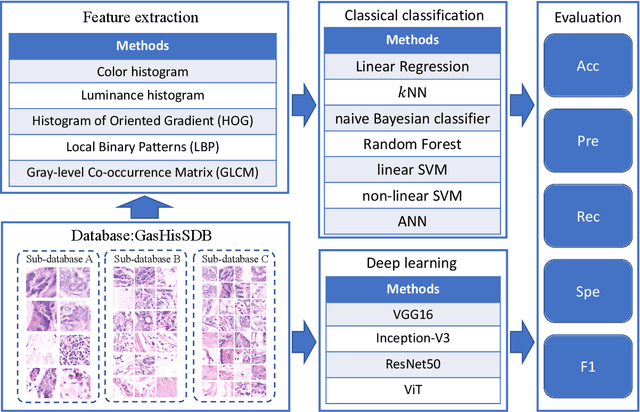
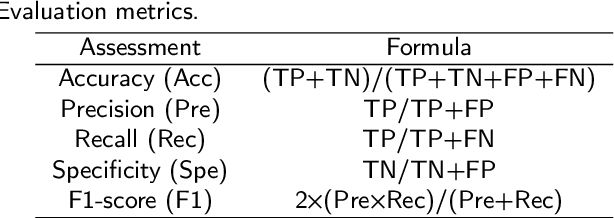
Gastric cancer is the fifth most common cancer in the world. At the same time, it is also the fourth most deadly cancer. Early detection of cancer exists as a guide for the treatment of gastric cancer. Nowadays, computer technology has advanced rapidly to assist physicians in the diagnosis of pathological pictures of gastric cancer. Ensemble learning is a way to improve the accuracy of algorithms, and finding multiple learning models with complementarity types is the basis of ensemble learning. The complementarity of sub-size pathology image classifiers when machine performance is insufficient is explored in this experimental platform. We choose seven classical machine learning classifiers and four deep learning classifiers for classification experiments on the GasHisSDB database. Among them, classical machine learning algorithms extract five different image virtual features to match multiple classifier algorithms. For deep learning, we choose three convolutional neural network classifiers. In addition, we also choose a novel Transformer-based classifier. The experimental platform, in which a large number of classical machine learning and deep learning methods are performed, demonstrates that there are differences in the performance of different classifiers on GasHisSDB. Classical machine learning models exist for classifiers that classify Abnormal categories very well, while classifiers that excel in classifying Normal categories also exist. Deep learning models also exist with multiple models that can be complementarity. Suitable classifiers are selected for ensemble learning, when machine performance is insufficient. This experimental platform demonstrates that multiple classifiers are indeed complementarity and can improve the efficiency of ensemble learning. This can better assist doctors in diagnosis, improve the detection of gastric cancer, and increase the cure rate.
Discovery Radiomics via Deep Multi-Column Radiomic Sequencers for Skin Cancer Detection
Sep 24, 2017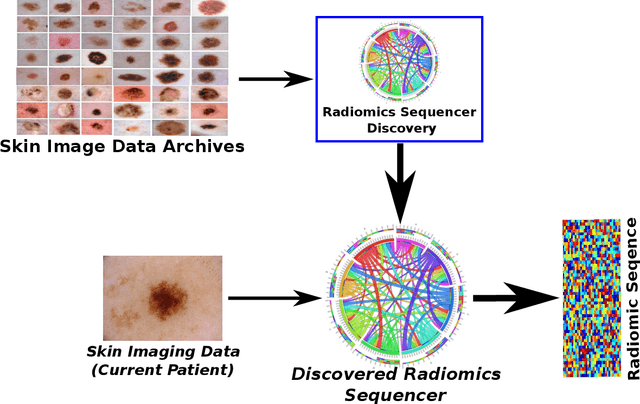
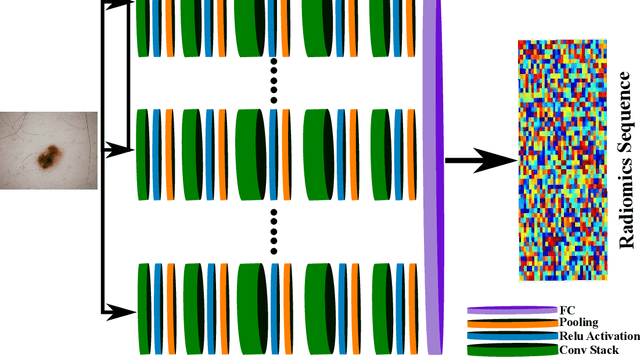
While skin cancer is the most diagnosed form of cancer in men and women, with more cases diagnosed each year than all other cancers combined, sufficiently early diagnosis results in very good prognosis and as such makes early detection crucial. While radiomics have shown considerable promise as a powerful diagnostic tool for significantly improving oncological diagnostic accuracy and efficiency, current radiomics-driven methods have largely rely on pre-defined, hand-crafted quantitative features, which can greatly limit the ability to fully characterize unique cancer phenotype that distinguish it from healthy tissue. Recently, the notion of discovery radiomics was introduced, where a large amount of custom, quantitative radiomic features are directly discovered from the wealth of readily available medical imaging data. In this study, we present a novel discovery radiomics framework for skin cancer detection, where we leverage novel deep multi-column radiomic sequencers for high-throughput discovery and extraction of a large amount of custom radiomic features tailored for characterizing unique skin cancer tissue phenotype. The discovered radiomic sequencer was tested against 9,152 biopsy-proven clinical images comprising of different skin cancers such as melanoma and basal cell carcinoma, and demonstrated sensitivity and specificity of 91% and 75%, respectively, thus achieving dermatologist-level performance and \break hence can be a powerful tool for assisting general practitioners and dermatologists alike in improving the efficiency, consistency, and accuracy of skin cancer diagnosis.
Towards Confident Detection of Prostate Cancer using High Resolution Micro-ultrasound
Jul 21, 2022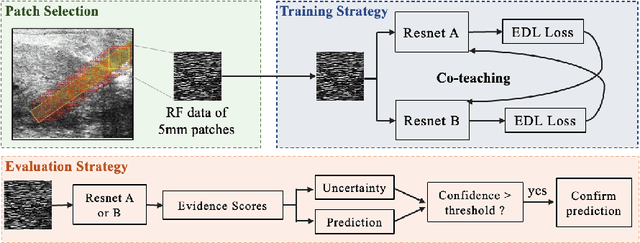
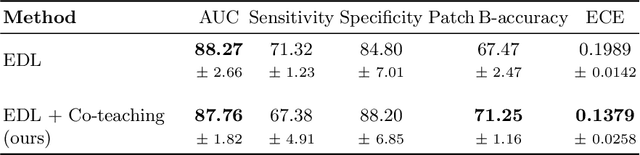
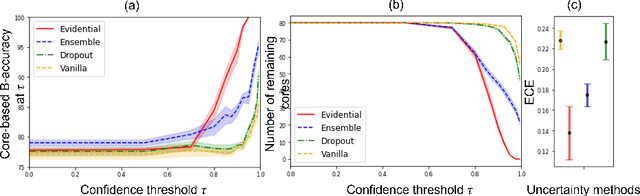
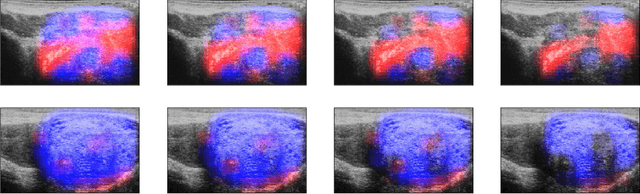
MOTIVATION: Detection of prostate cancer during transrectal ultrasound-guided biopsy is challenging. The highly heterogeneous appearance of cancer, presence of ultrasound artefacts, and noise all contribute to these difficulties. Recent advancements in high-frequency ultrasound imaging - micro-ultrasound - have drastically increased the capability of tissue imaging at high resolution. Our aim is to investigate the development of a robust deep learning model specifically for micro-ultrasound-guided prostate cancer biopsy. For the model to be clinically adopted, a key challenge is to design a solution that can confidently identify the cancer, while learning from coarse histopathology measurements of biopsy samples that introduce weak labels. METHODS: We use a dataset of micro-ultrasound images acquired from 194 patients, who underwent prostate biopsy. We train a deep model using a co-teaching paradigm to handle noise in labels, together with an evidential deep learning method for uncertainty estimation. We evaluate the performance of our model using the clinically relevant metric of accuracy vs. confidence. RESULTS: Our model achieves a well-calibrated estimation of predictive uncertainty with area under the curve of 88$\%$. The use of co-teaching and evidential deep learning in combination yields significantly better uncertainty estimation than either alone. We also provide a detailed comparison against state-of-the-art in uncertainty estimation.
Cell nuclei classification in histopathological images using hybrid OLConvNet
Feb 21, 2022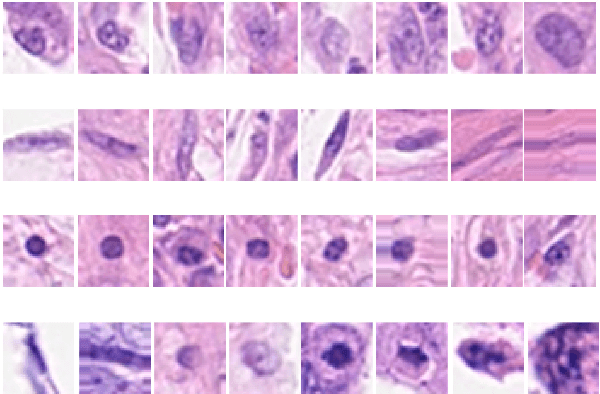
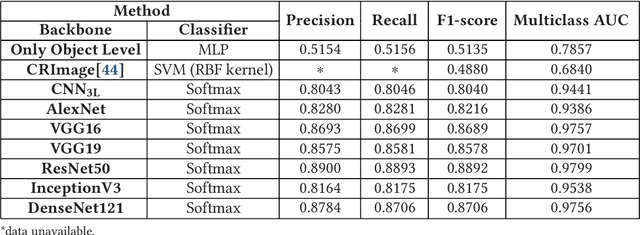
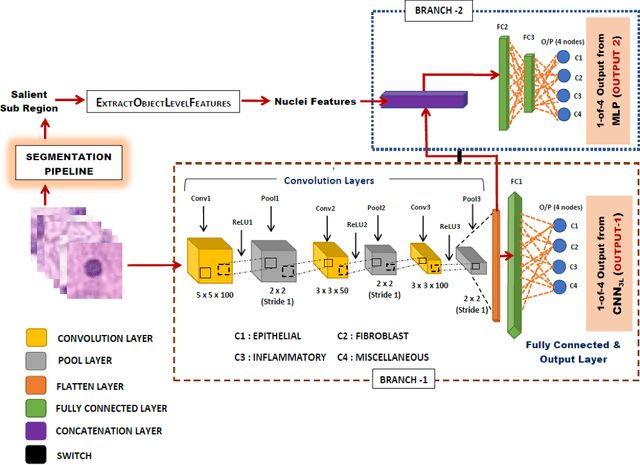
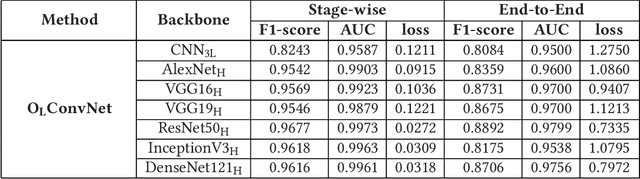
Computer-aided histopathological image analysis for cancer detection is a major research challenge in the medical domain. Automatic detection and classification of nuclei for cancer diagnosis impose a lot of challenges in developing state of the art algorithms due to the heterogeneity of cell nuclei and data set variability. Recently, a multitude of classification algorithms has used complex deep learning models for their dataset. However, most of these methods are rigid and their architectural arrangement suffers from inflexibility and non-interpretability. In this research article, we have proposed a hybrid and flexible deep learning architecture OLConvNet that integrates the interpretability of traditional object-level features and generalization of deep learning features by using a shallower Convolutional Neural Network (CNN) named as $CNN_{3L}$. $CNN_{3L}$ reduces the training time by training fewer parameters and hence eliminating space constraints imposed by deeper algorithms. We used F1-score and multiclass Area Under the Curve (AUC) performance parameters to compare the results. To further strengthen the viability of our architectural approach, we tested our proposed methodology with state of the art deep learning architectures AlexNet, VGG16, VGG19, ResNet50, InceptionV3, and DenseNet121 as backbone networks. After a comprehensive analysis of classification results from all four architectures, we observed that our proposed model works well and perform better than contemporary complex algorithms.
Region of Interest Detection in Melanocytic Skin Tumor Whole Slide Images
Oct 29, 2022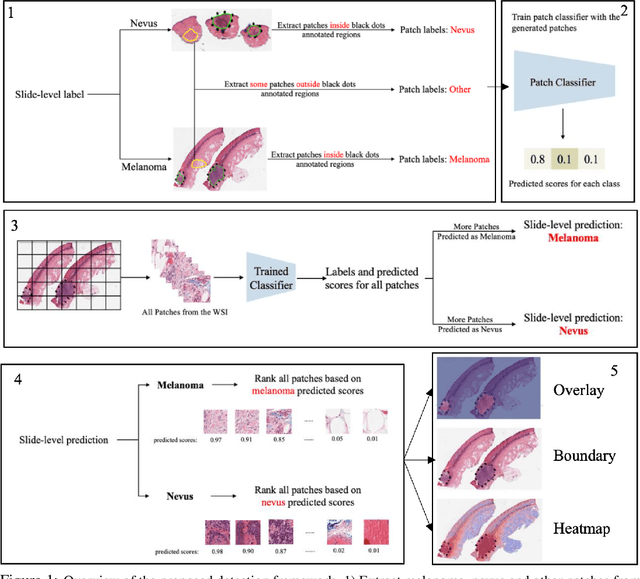
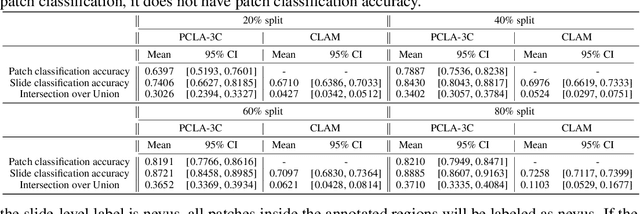
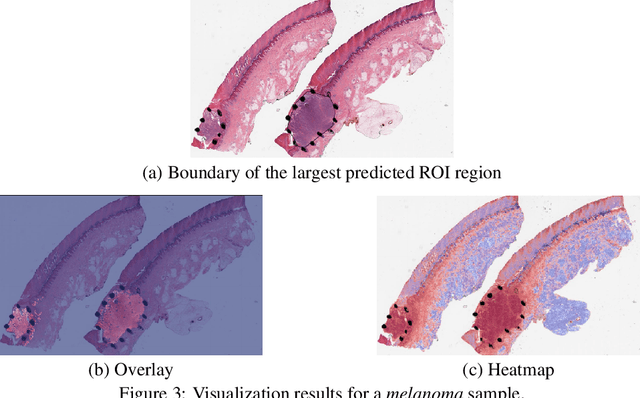
Automated region of interest detection in histopathological image analysis is a challenging and important topic with tremendous potential impact on clinical practice. The deep-learning methods used in computational pathology help us to reduce costs and increase the speed and accuracy of regions of interest detection and cancer diagnosis. In this work, we propose a patch-based region of interest detection method for melanocytic skin tumor whole-slide images. We work with a dataset that contains 165 primary melanomas and nevi Hematoxylin and Eosin whole-slide images and build a deep-learning method. The proposed method performs well on a hold-out test data set including five TCGA-SKCM slides (accuracy of 93.94\% in slide classification task and intersection over union rate of 41.27\% in the region of interest detection task), showing the outstanding performance of our model on melanocytic skin tumor. Even though we test the experiments on the skin tumor dataset, our work could also be extended to other medical image detection problems, such as various tumors' classification and prediction, to help and benefit the clinical evaluation and diagnosis of different tumors.
Predicting Real-time Scientific Experiments Using Transformer models and Reinforcement Learning
Apr 25, 2022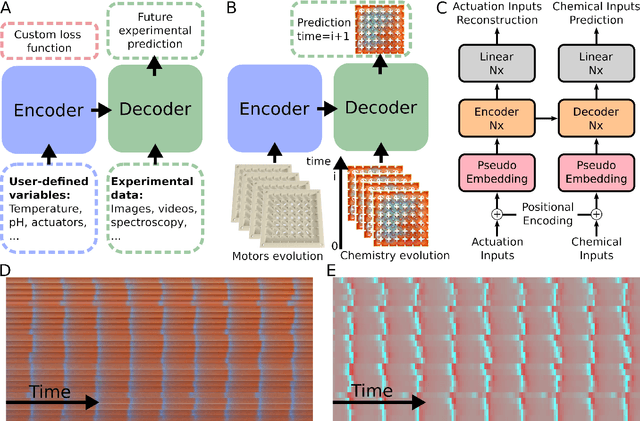
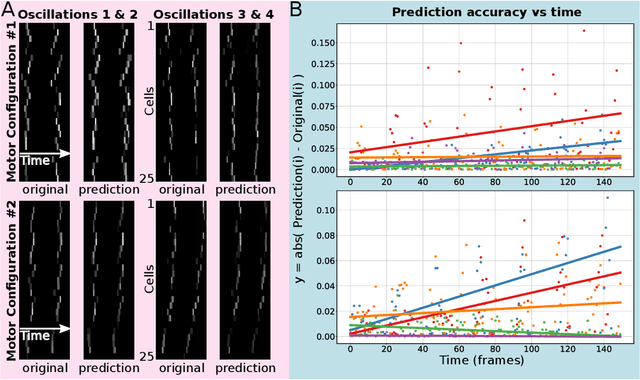
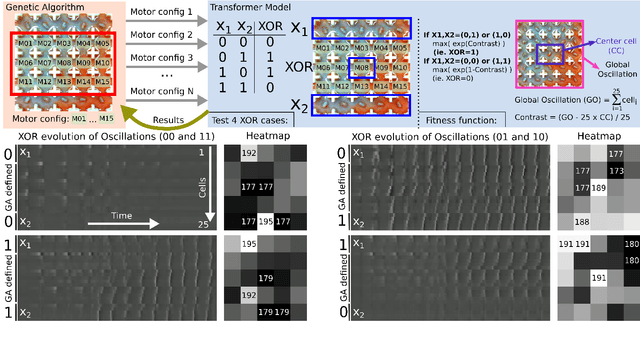
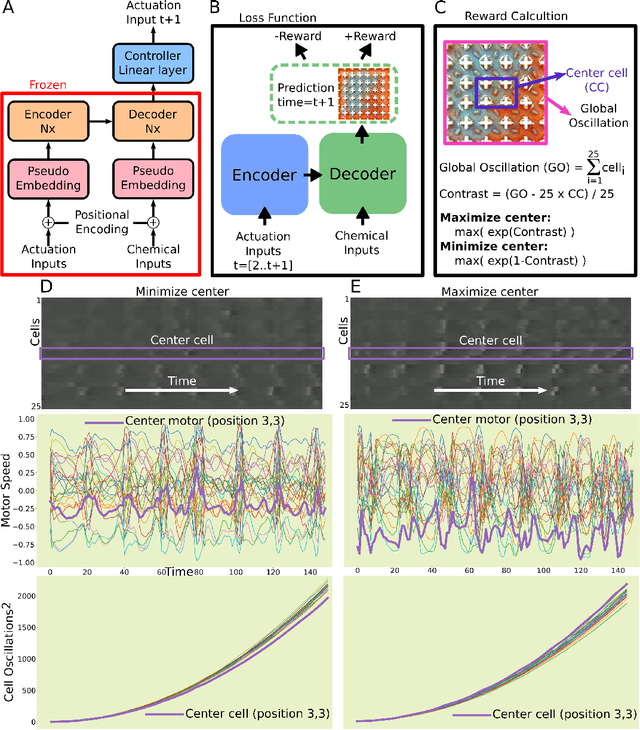
Life and physical sciences have always been quick to adopt the latest advances in machine learning to accelerate scientific discovery. Examples of this are cell segmentation or cancer detection. Nevertheless, these exceptional results are based on mining previously created datasets to discover patterns or trends. Recent advances in AI have been demonstrated in real-time scenarios like self-driving cars or playing video games. However, these new techniques have not seen widespread adoption in life or physical sciences because experimentation can be slow. To tackle this limitation, this work aims to adapt generative learning algorithms to model scientific experiments and accelerate their discovery using in-silico simulations. We particularly focused on real-time experiments, aiming to model how they react to user inputs. To achieve this, here we present an encoder-decoder architecture based on the Transformer model to simulate real-time scientific experimentation, predict its future behaviour and manipulate it on a step-by-step basis. As a proof of concept, this architecture was trained to map a set of mechanical inputs to the oscillations generated by a chemical reaction. The model was paired with a Reinforcement Learning controller to show how the simulated chemistry can be manipulated in real-time towards user-defined behaviours. Our results demonstrate how generative learning can model real-time scientific experimentation to track how it changes through time as the user manipulates it, and how the trained models can be paired with optimisation algorithms to discover new phenomena beyond the physical limitations of lab experimentation. This work paves the way towards building surrogate systems where physical experimentation interacts with machine learning on a step-by-step basis.
* 8 pages, 5 figures, conference
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Scalable privacy-preserving cancer type prediction with homomorphic encryption
Apr 12, 2022
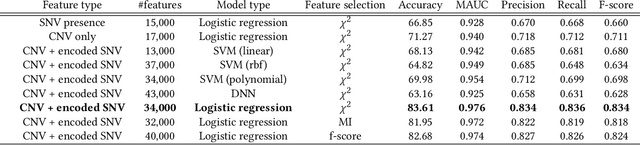
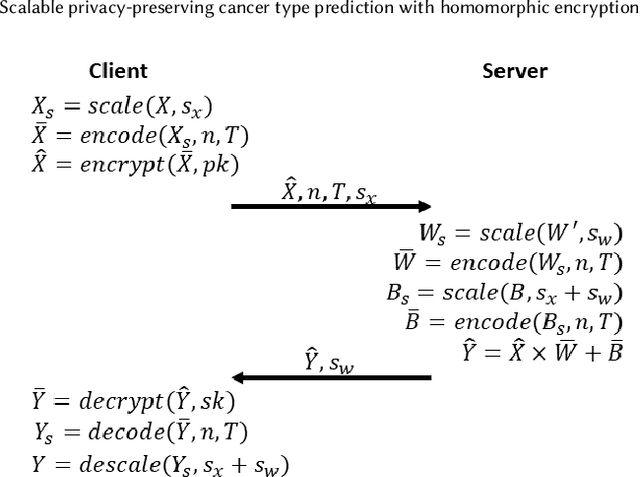
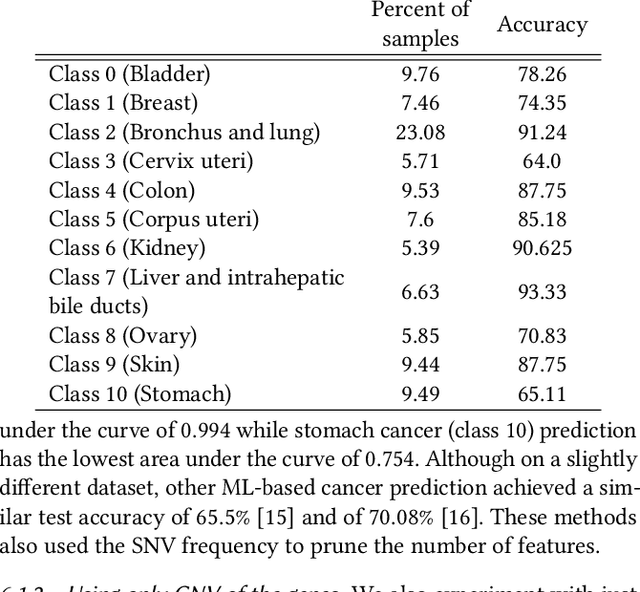
Machine Learning (ML) alleviates the challenges of high-dimensional data analysis and improves decision making in critical applications like healthcare. Effective cancer type from high-dimensional genetic mutation data can be useful for cancer diagnosis and treatment, if the distinguishable patterns between cancer types are identified. At the same time, analysis of high-dimensional data is computationally expensive and is often outsourced to cloud services. Privacy concerns in outsourced ML, especially in the field of genetics, motivate the use of encrypted computation, like Homomorphic Encryption (HE). But restrictive overheads of encrypted computation deter its usage. In this work, we explore the challenges of privacy preserving cancer detection using a real-world dataset consisting of more than 2 million genetic information for several cancer types. Since the data is inherently high-dimensional, we explore smaller ML models for cancer prediction to enable fast inference in the privacy preserving domain. We develop a solution for privacy preserving cancer inference which first leverages the domain knowledge on somatic mutations to efficiently encode genetic mutations and then uses statistical tests for feature selection. Our logistic regression model, built using our novel encoding scheme, achieves 0.98 micro-average area under curve with 13% higher test accuracy than similar studies. We exhaustively test our model's predictive capabilities by analyzing the genes used by the model. Furthermore, we propose a fast matrix multiplication algorithm that can efficiently handle high-dimensional data. Experimental results show that, even with 40,000 features, our proposed matrix multiplication algorithm can speed up concurrent inference of multiple individuals by approximately 10x and inference of a single individual by approximately 550x, in comparison to standard matrix multiplication.
Pit-Pattern Classification of Colorectal Cancer Polyps Using a Hyper Sensitive Vision-Based Tactile Sensor and Dilated Residual Networks
Nov 13, 2022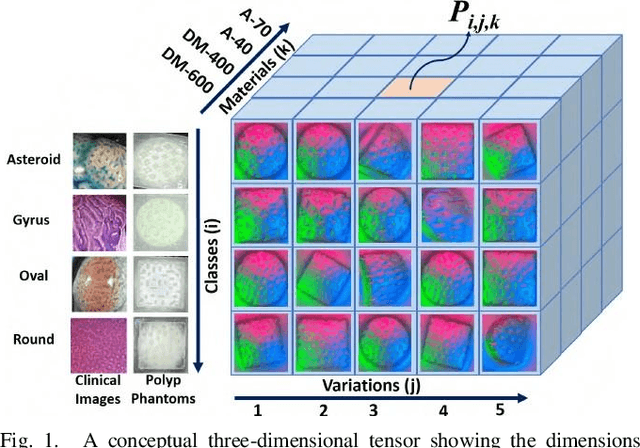
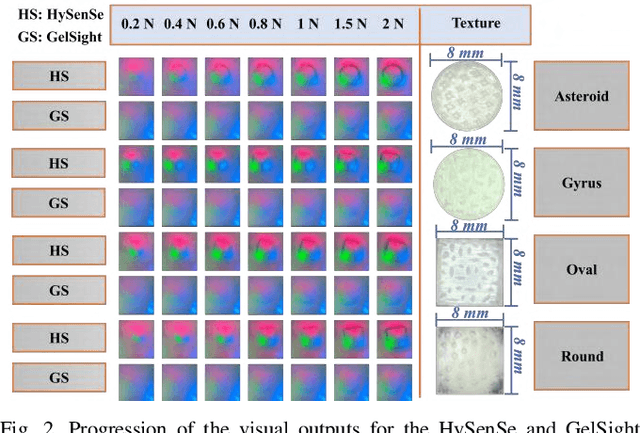
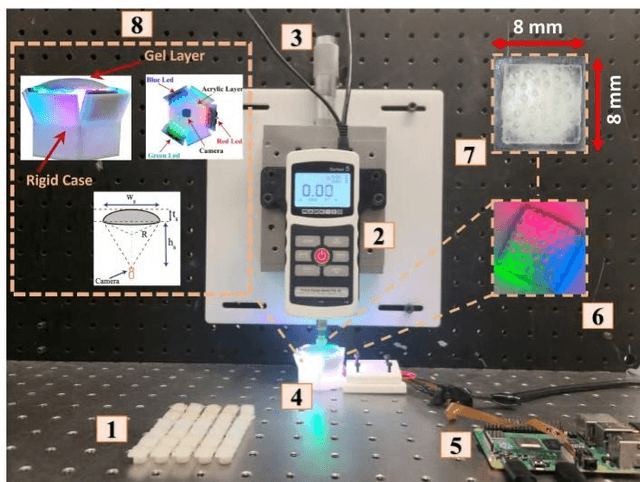
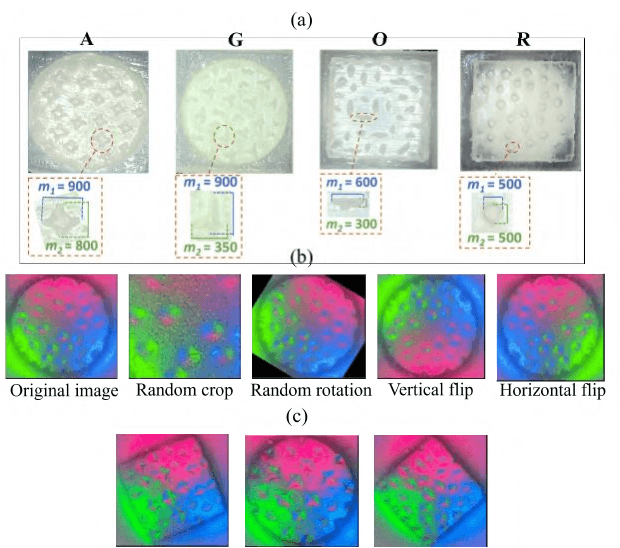
In this study, with the goal of reducing the early detection miss rate of colorectal cancer (CRC) polyps, we propose utilizing a novel hyper-sensitive vision-based tactile sensor called HySenSe and a complementary and novel machine learning (ML) architecture that explores the potentials of utilizing dilated convolutions, the beneficial features of the ResNet architecture, and the transfer learning concept applied on a small dataset with the scale of hundreds of images. The proposed tactile sensor provides high-resolution 3D textural images of CRC polyps that will be used for their accurate classification via the proposed dilated residual network. To collect realistic surface patterns of CRC polyps for training the ML models and evaluating their performance, we first designed and additively manufactured 160 unique realistic polyp phantoms consisting of 4 different hardness. Next, the proposed architecture was compared with the state-of-the-art ML models (e.g., AlexNet and DenseNet) and proved to be superior in terms of performance and complexity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge