Benedikt Wiestler
for the ALFA study
Optimizing Rank for High-Fidelity Implicit Neural Representations
Dec 16, 2025Abstract:Implicit Neural Representations (INRs) based on vanilla Multi-Layer Perceptrons (MLPs) are widely believed to be incapable of representing high-frequency content. This has directed research efforts towards architectural interventions, such as coordinate embeddings or specialized activation functions, to represent high-frequency signals. In this paper, we challenge the notion that the low-frequency bias of vanilla MLPs is an intrinsic, architectural limitation to learn high-frequency content, but instead a symptom of stable rank degradation during training. We empirically demonstrate that regulating the network's rank during training substantially improves the fidelity of the learned signal, rendering even simple MLP architectures expressive. Extensive experiments show that using optimizers like Muon, with high-rank, near-orthogonal updates, consistently enhances INR architectures even beyond simple ReLU MLPs. These substantial improvements hold across a diverse range of domains, including natural and medical images, and novel view synthesis, with up to 9 dB PSNR improvements over the previous state-of-the-art. Our project page, which includes code and experimental results, is available at: (https://muon-inrs.github.io).
Learning to reason about rare diseases through retrieval-augmented agents
Nov 06, 2025Abstract:Rare diseases represent the long tail of medical imaging, where AI models often fail due to the scarcity of representative training data. In clinical workflows, radiologists frequently consult case reports and literature when confronted with unfamiliar findings. Following this line of reasoning, we introduce RADAR, Retrieval Augmented Diagnostic Reasoning Agents, an agentic system for rare disease detection in brain MRI. Our approach uses AI agents with access to external medical knowledge by embedding both case reports and literature using sentence transformers and indexing them with FAISS to enable efficient similarity search. The agent retrieves clinically relevant evidence to guide diagnostic decision making on unseen diseases, without the need of additional training. Designed as a model-agnostic reasoning module, RADAR can be seamlessly integrated with diverse large language models, consistently improving their rare pathology recognition and interpretability. On the NOVA dataset comprising 280 distinct rare diseases, RADAR achieves up to a 10.2% performance gain, with the strongest improvements observed for open source models such as DeepSeek. Beyond accuracy, the retrieved examples provide interpretable, literature grounded explanations, highlighting retrieval-augmented reasoning as a powerful paradigm for low-prevalence conditions in medical imaging.
BioSonix: Can Physics-Based Sonification Perceptualize Tissue Deformations From Tool Interactions?
Aug 20, 2025Abstract:Perceptualizing tool interactions with deformable structures in surgical procedures remains challenging, as unimodal visualization techniques often fail to capture the complexity of these interactions due to constraints such as occlusion and limited depth perception. This paper presents a novel approach to augment tool navigation in mixed reality environments by providing auditory representations of tool-tissue dynamics, particularly for interactions with soft tissue. BioSonix, a physics-informed design framework, utilizes tissue displacements in 3D space to compute excitation forces for a sound model encoding tissue properties such as stiffness and density. Biomechanical simulations were employed to model particle displacements resulting from tool-tissue interactions, establishing a robust foundation for the method. An optimization approach was used to define configurations for capturing diverse interaction scenarios with varying tool trajectories. Experiments were conducted to validate the accuracy of the sound-displacement mappings. Additionally, two user studies were performed: the first involved two clinical professionals (a neuroradiologist and a cardiologist), who confirmed the method's impact and achieved high task accuracy; the second included 22 biomedical experts, who demonstrated high discrimination accuracy in tissue differentiation and targeting tasks. The results revealed a strong correlation between tool-tissue dynamics and their corresponding auditory profiles, highlighting the potential of these sound representations to enhance the intuitive understanding of complex interactions.
* V. Ruozzi and S. Matinfar contributed equally to this work
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025



Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
NOVA: A Benchmark for Anomaly Localization and Clinical Reasoning in Brain MRI
May 20, 2025



Abstract:In many real-world applications, deployed models encounter inputs that differ from the data seen during training. Out-of-distribution detection identifies whether an input stems from an unseen distribution, while open-world recognition flags such inputs to ensure the system remains robust as ever-emerging, previously $unknown$ categories appear and must be addressed without retraining. Foundation and vision-language models are pre-trained on large and diverse datasets with the expectation of broad generalization across domains, including medical imaging. However, benchmarking these models on test sets with only a few common outlier types silently collapses the evaluation back to a closed-set problem, masking failures on rare or truly novel conditions encountered in clinical use. We therefore present $NOVA$, a challenging, real-life $evaluation-only$ benchmark of $\sim$900 brain MRI scans that span 281 rare pathologies and heterogeneous acquisition protocols. Each case includes rich clinical narratives and double-blinded expert bounding-box annotations. Together, these enable joint assessment of anomaly localisation, visual captioning, and diagnostic reasoning. Because NOVA is never used for training, it serves as an $extreme$ stress-test of out-of-distribution generalisation: models must bridge a distribution gap both in sample appearance and in semantic space. Baseline results with leading vision-language models (GPT-4o, Gemini 2.0 Flash, and Qwen2.5-VL-72B) reveal substantial performance drops across all tasks, establishing NOVA as a rigorous testbed for advancing models that can detect, localize, and reason about truly unknown anomalies.
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
TimeFlow: Longitudinal Brain Image Registration and Aging Progression Analysis
Jan 15, 2025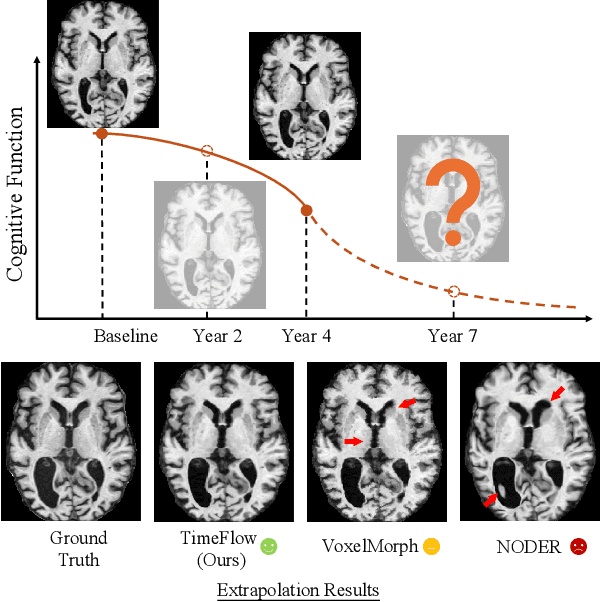


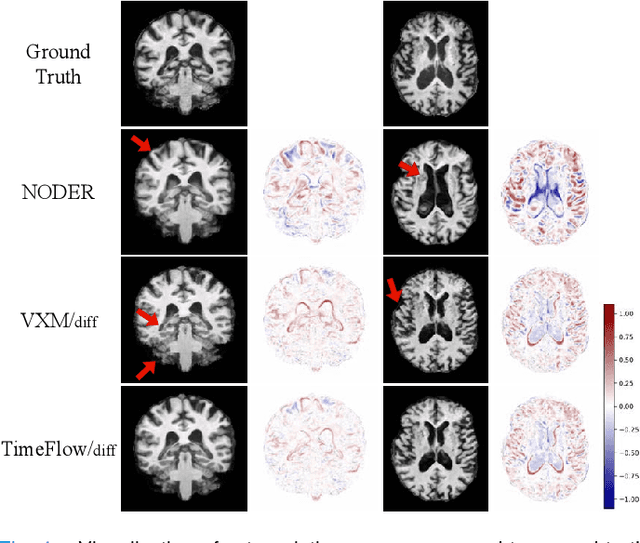
Abstract:Predicting future brain states is crucial for understanding healthy aging and neurodegenerative diseases. Longitudinal brain MRI registration, a cornerstone for such analyses, has long been limited by its inability to forecast future developments, reliance on extensive, dense longitudinal data, and the need to balance registration accuracy with temporal smoothness. In this work, we present \emph{TimeFlow}, a novel framework for longitudinal brain MRI registration that overcomes all these challenges. Leveraging a U-Net architecture with temporal conditioning inspired by diffusion models, TimeFlow enables accurate longitudinal registration and facilitates prospective analyses through future image prediction. Unlike traditional methods that depend on explicit smoothness regularizers and dense sequential data, TimeFlow achieves temporal consistency and continuity without these constraints. Experimental results highlight its superior performance in both future timepoint prediction and registration accuracy compared to state-of-the-art methods. Additionally, TimeFlow supports novel biological brain aging analyses, effectively differentiating neurodegenerative conditions from healthy aging. It eliminates the need for segmentation, thereby avoiding the challenges of non-trivial annotation and inconsistent segmentation errors. TimeFlow paves the way for accurate, data-efficient, and annotation-free prospective analyses of brain aging and chronic diseases.
Efficient Deep Learning-based Forward Solvers for Brain Tumor Growth Models
Jan 14, 2025Abstract:Glioblastoma, a highly aggressive brain tumor, poses major challenges due to its poor prognosis and high morbidity rates. Partial differential equation-based models offer promising potential to enhance therapeutic outcomes by simulating patient-specific tumor behavior for improved radiotherapy planning. However, model calibration remains a bottleneck due to the high computational demands of optimization methods like Monte Carlo sampling and evolutionary algorithms. To address this, we recently introduced an approach leveraging a neural forward solver with gradient-based optimization to significantly reduce calibration time. This approach requires a highly accurate and fully differentiable forward model. We investigate multiple architectures, including (i) an enhanced TumorSurrogate, (ii) a modified nnU-Net, and (iii) a 3D Vision Transformer (ViT). The optimized TumorSurrogate achieved the best overall results, excelling in both tumor outline matching and voxel-level prediction of tumor cell concentration. It halved the MSE relative to the baseline model and achieved the highest Dice score across all tumor cell concentration thresholds. Our study demonstrates significant enhancement in forward solver performance and outlines important future research directions.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Spatial Brain Tumor Concentration Estimation for Individualized Radiotherapy Planning
Dec 18, 2024



Abstract:Biophysical modeling of brain tumors has emerged as a promising strategy for personalizing radiotherapy planning by estimating the otherwise hidden distribution of tumor cells within the brain. However, many existing state-of-the-art methods are computationally intensive, limiting their widespread translation into clinical practice. In this work, we propose an efficient and direct method that utilizes soft physical constraints to estimate the tumor cell concentration from preoperative MRI of brain tumor patients. Our approach optimizes a 3D tumor concentration field by simultaneously minimizing the difference between the observed MRI and a physically informed loss function. Compared to existing state-of-the-art techniques, our method significantly improves predicting tumor recurrence on two public datasets with a total of 192 patients while maintaining a clinically viable runtime of under one minute - a substantial reduction from the 30 minutes required by the current best approach. Furthermore, we showcase the generalizability of our framework by incorporating additional imaging information and physical constraints, highlighting its potential to translate to various medical diffusion phenomena with imperfect data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge