Prateek Prasanna
TICON: A Slide-Level Tile Contextualizer for Histopathology Representation Learning
Dec 24, 2025Abstract:The interpretation of small tiles in large whole slide images (WSI) often needs a larger image context. We introduce TICON, a transformer-based tile representation contextualizer that produces rich, contextualized embeddings for ''any'' application in computational pathology. Standard tile encoder-based pipelines, which extract embeddings of tiles stripped from their context, fail to model the rich slide-level information essential for both local and global tasks. Furthermore, different tile-encoders excel at different downstream tasks. Therefore, a unified model is needed to contextualize embeddings derived from ''any'' tile-level foundation model. TICON addresses this need with a single, shared encoder, pretrained using a masked modeling objective to simultaneously unify and contextualize representations from diverse tile-level pathology foundation models. Our experiments demonstrate that TICON-contextualized embeddings significantly improve performance across many different tasks, establishing new state-of-the-art results on tile-level benchmarks (i.e., HEST-Bench, THUNDER, CATCH) and slide-level benchmarks (i.e., Patho-Bench). Finally, we pretrain an aggregator on TICON to form a slide-level foundation model, using only 11K WSIs, outperforming SoTA slide-level foundation models pretrained with up to 350K WSIs.
NeuroRAD-FM: A Foundation Model for Neuro-Oncology with Distributionally Robust Training
Sep 18, 2025Abstract:Neuro-oncology poses unique challenges for machine learning due to heterogeneous data and tumor complexity, limiting the ability of foundation models (FMs) to generalize across cohorts. Existing FMs also perform poorly in predicting uncommon molecular markers, which are essential for treatment response and risk stratification. To address these gaps, we developed a neuro-oncology specific FM with a distributionally robust loss function, enabling accurate estimation of tumor phenotypes while maintaining cross-institution generalization. We pretrained self-supervised backbones (BYOL, DINO, MAE, MoCo) on multi-institutional brain tumor MRI and applied distributionally robust optimization (DRO) to mitigate site and class imbalance. Downstream tasks included molecular classification of common markers (MGMT, IDH1, 1p/19q, EGFR), uncommon alterations (ATRX, TP53, CDKN2A/2B, TERT), continuous markers (Ki-67, TP53), and overall survival prediction in IDH1 wild-type glioblastoma at UCSF, UPenn, and CUIMC. Our method improved molecular prediction and reduced site-specific embedding differences. At CUIMC, mean balanced accuracy rose from 0.744 to 0.785 and AUC from 0.656 to 0.676, with the largest gains for underrepresented endpoints (CDKN2A/2B accuracy 0.86 to 0.92, AUC 0.73 to 0.92; ATRX AUC 0.69 to 0.82; Ki-67 accuracy 0.60 to 0.69). For survival, c-index improved at all sites: CUIMC 0.592 to 0.597, UPenn 0.647 to 0.672, UCSF 0.600 to 0.627. Grad-CAM highlighted tumor and peri-tumoral regions, confirming interpretability. Overall, coupling FMs with DRO yields more site-invariant representations, improves prediction of common and uncommon markers, and enhances survival discrimination, underscoring the need for prospective validation and integration of longitudinal and interventional signals to advance precision neuro-oncology.
PixCell: A generative foundation model for digital histopathology images
Jun 05, 2025Abstract:The digitization of histology slides has revolutionized pathology, providing massive datasets for cancer diagnosis and research. Contrastive self-supervised and vision-language models have been shown to effectively mine large pathology datasets to learn discriminative representations. On the other hand, generative models, capable of synthesizing realistic and diverse images, present a compelling solution to address unique problems in pathology that involve synthesizing images; overcoming annotated data scarcity, enabling privacy-preserving data sharing, and performing inherently generative tasks, such as virtual staining. We introduce PixCell, the first diffusion-based generative foundation model for histopathology. We train PixCell on PanCan-30M, a vast, diverse dataset derived from 69,184 H\&E-stained whole slide images covering various cancer types. We employ a progressive training strategy and a self-supervision-based conditioning that allows us to scale up training without any annotated data. PixCell generates diverse and high-quality images across multiple cancer types, which we find can be used in place of real data to train a self-supervised discriminative model. Synthetic images shared between institutions are subject to fewer regulatory barriers than would be the case with real clinical images. Furthermore, we showcase the ability to precisely control image generation using a small set of annotated images, which can be used for both data augmentation and educational purposes. Testing on a cell segmentation task, a mask-guided PixCell enables targeted data augmentation, improving downstream performance. Finally, we demonstrate PixCell's ability to use H\&E structural staining to infer results from molecular marker studies; we use this capability to infer IHC staining from H\&E images. Our trained models are publicly released to accelerate research in computational pathology.
ImmunoDiff: A Diffusion Model for Immunotherapy Response Prediction in Lung Cancer
May 29, 2025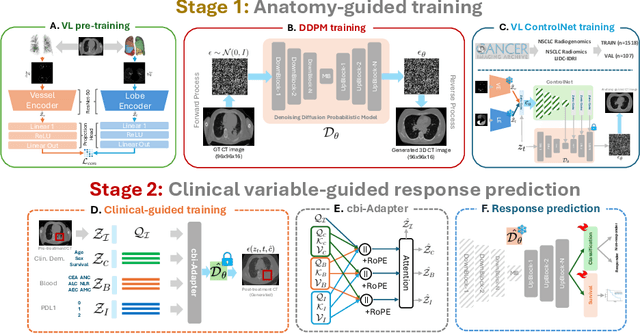

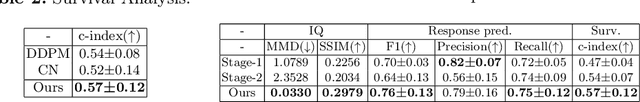

Abstract:Accurately predicting immunotherapy response in Non-Small Cell Lung Cancer (NSCLC) remains a critical unmet need. Existing radiomics and deep learning-based predictive models rely primarily on pre-treatment imaging to predict categorical response outcomes, limiting their ability to capture the complex morphological and textural transformations induced by immunotherapy. This study introduces ImmunoDiff, an anatomy-aware diffusion model designed to synthesize post-treatment CT scans from baseline imaging while incorporating clinically relevant constraints. The proposed framework integrates anatomical priors, specifically lobar and vascular structures, to enhance fidelity in CT synthesis. Additionally, we introduce a novel cbi-Adapter, a conditioning module that ensures pairwise-consistent multimodal integration of imaging and clinical data embeddings, to refine the generative process. Additionally, a clinical variable conditioning mechanism is introduced, leveraging demographic data, blood-based biomarkers, and PD-L1 expression to refine the generative process. Evaluations on an in-house NSCLC cohort treated with immune checkpoint inhibitors demonstrate a 21.24% improvement in balanced accuracy for response prediction and a 0.03 increase in c-index for survival prediction. Code will be released soon.
BrainMRDiff: A Diffusion Model for Anatomically Consistent Brain MRI Synthesis
Apr 06, 2025



Abstract:Accurate brain tumor diagnosis relies on the assessment of multiple Magnetic Resonance Imaging (MRI) sequences. However, in clinical practice, the acquisition of certain sequences may be affected by factors like motion artifacts or contrast agent contraindications, leading to suboptimal outcome, such as poor image quality. This can then affect image interpretation by radiologists. Synthesizing high quality MRI sequences has thus become a critical research focus. Though recent advancements in controllable generative AI have facilitated the synthesis of diagnostic quality MRI, ensuring anatomical accuracy remains a significant challenge. Preserving critical structural relationships between different anatomical regions is essential, as even minor structural or topological inconsistencies can compromise diagnostic validity. In this work, we propose BrainMRDiff, a novel topology-preserving, anatomy-guided diffusion model for synthesizing brain MRI, leveraging brain and tumor anatomies as conditioning inputs. To achieve this, we introduce two key modules: Tumor+Structure Aggregation (TSA) and Topology-Guided Anatomy Preservation (TGAP). TSA integrates diverse anatomical structures with tumor information, forming a comprehensive conditioning mechanism for the diffusion process. TGAP enforces topological consistency during reverse denoising diffusion process; both these modules ensure that the generated image respects anatomical integrity. Experimental results demonstrate that BrainMRDiff surpasses existing baselines, achieving performance improvements of 23.33% on the BraTS-AG dataset and 33.33% on the BraTS-Met dataset. Code will be made publicly available soon.
GECKO: Gigapixel Vision-Concept Contrastive Pretraining in Histopathology
Apr 01, 2025Abstract:Pretraining a Multiple Instance Learning (MIL) aggregator enables the derivation of Whole Slide Image (WSI)-level embeddings from patch-level representations without supervision. While recent multimodal MIL pretraining approaches leveraging auxiliary modalities have demonstrated performance gains over unimodal WSI pretraining, the acquisition of these additional modalities necessitates extensive clinical profiling. This requirement increases costs and limits scalability in existing WSI datasets lacking such paired modalities. To address this, we propose Gigapixel Vision-Concept Knowledge Contrastive pretraining (GECKO), which aligns WSIs with a Concept Prior derived from the available WSIs. First, we derive an inherently interpretable concept prior by computing the similarity between each WSI patch and textual descriptions of predefined pathology concepts. GECKO then employs a dual-branch MIL network: one branch aggregates patch embeddings into a WSI-level deep embedding, while the other aggregates the concept prior into a corresponding WSI-level concept embedding. Both aggregated embeddings are aligned using a contrastive objective, thereby pretraining the entire dual-branch MIL model. Moreover, when auxiliary modalities such as transcriptomics data are available, GECKO seamlessly integrates them. Across five diverse tasks, GECKO consistently outperforms prior unimodal and multimodal pretraining approaches while also delivering clinically meaningful interpretability that bridges the gap between computational models and pathology expertise. Code is made available at https://github.com/bmi-imaginelab/GECKO
Pathology Image Compression with Pre-trained Autoencoders
Mar 14, 2025Abstract:The growing volume of high-resolution Whole Slide Images in digital histopathology poses significant storage, transmission, and computational efficiency challenges. Standard compression methods, such as JPEG, reduce file sizes but often fail to preserve fine-grained phenotypic details critical for downstream tasks. In this work, we repurpose autoencoders (AEs) designed for Latent Diffusion Models as an efficient learned compression framework for pathology images. We systematically benchmark three AE models with varying compression levels and evaluate their reconstruction ability using pathology foundation models. We introduce a fine-tuning strategy to further enhance reconstruction fidelity that optimizes a pathology-specific learned perceptual metric. We validate our approach on downstream tasks, including segmentation, patch classification, and multiple instance learning, showing that replacing images with AE-compressed reconstructions leads to minimal performance degradation. Additionally, we propose a K-means clustering-based quantization method for AE latents, improving storage efficiency while maintaining reconstruction quality. We provide the weights of the fine-tuned autoencoders at https://huggingface.co/collections/StonyBrook-CVLab/pathology-fine-tuned-aes-67d45f223a659ff2e3402dd0.
Enhancing SAM with Efficient Prompting and Preference Optimization for Semi-supervised Medical Image Segmentation
Mar 06, 2025



Abstract:Foundational models such as the Segment Anything Model (SAM) are gaining traction in medical imaging segmentation, supporting multiple downstream tasks. However, such models are supervised in nature, still relying on large annotated datasets or prompts supplied by experts. Conventional techniques such as active learning to alleviate such limitations are limited in scope and still necessitate continuous human involvement and complex domain knowledge for label refinement or establishing reward ground truth. To address these challenges, we propose an enhanced Segment Anything Model (SAM) framework that utilizes annotation-efficient prompts generated in a fully unsupervised fashion, while still capturing essential semantic, location, and shape information through contrastive language-image pretraining and visual question answering. We adopt the direct preference optimization technique to design an optimal policy that enables the model to generate high-fidelity segmentations with simple ratings or rankings provided by a virtual annotator simulating the human annotation process. State-of-the-art performance of our framework in tasks such as lung segmentation, breast tumor segmentation, and organ segmentation across various modalities, including X-ray, ultrasound, and abdominal CT, justifies its effectiveness in low-annotation data scenarios.
TopoCellGen: Generating Histopathology Cell Topology with a Diffusion Model
Dec 08, 2024Abstract:Accurately modeling multi-class cell topology is crucial in digital pathology, as it provides critical insights into tissue structure and pathology. The synthetic generation of cell topology enables realistic simulations of complex tissue environments, enhances downstream tasks by augmenting training data, aligns more closely with pathologists' domain knowledge, and offers new opportunities for controlling and generalizing the tumor microenvironment. In this paper, we propose a novel approach that integrates topological constraints into a diffusion model to improve the generation of realistic, contextually accurate cell topologies. Our method refines the simulation of cell distributions and interactions, increasing the precision and interpretability of results in downstream tasks such as cell detection and classification. To assess the topological fidelity of generated layouts, we introduce a new metric, Topological Frechet Distance (TopoFD), which overcomes the limitations of traditional metrics like FID in evaluating topological structure. Experimental results demonstrate the effectiveness of our approach in generating multi-class cell layouts that capture intricate topological relationships.
Gen-SIS: Generative Self-augmentation Improves Self-supervised Learning
Dec 02, 2024



Abstract:Self-supervised learning (SSL) methods have emerged as strong visual representation learners by training an image encoder to maximize similarity between features of different views of the same image. To perform this view-invariance task, current SSL algorithms rely on hand-crafted augmentations such as random cropping and color jittering to create multiple views of an image. Recently, generative diffusion models have been shown to improve SSL by providing a wider range of data augmentations. However, these diffusion models require pre-training on large-scale image-text datasets, which might not be available for many specialized domains like histopathology. In this work, we introduce Gen-SIS, a diffusion-based augmentation technique trained exclusively on unlabeled image data, eliminating any reliance on external sources of supervision such as text captions. We first train an initial SSL encoder on a dataset using only hand-crafted augmentations. We then train a diffusion model conditioned on embeddings from that SSL encoder. Following training, given an embedding of the source image, this diffusion model can synthesize its diverse views. We show that these `self-augmentations', i.e. generative augmentations based on the vanilla SSL encoder embeddings, facilitate the training of a stronger SSL encoder. Furthermore, based on the ability to interpolate between images in the encoder latent space, we introduce the novel pretext task of disentangling the two source images of an interpolated synthetic image. We validate Gen-SIS's effectiveness by demonstrating performance improvements across various downstream tasks in both natural images, which are generally object-centric, as well as digital histopathology images, which are typically context-based.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge