Brian Wodlinger
ProstNFound+: A Prospective Study using Medical Foundation Models for Prostate Cancer Detection
Oct 30, 2025



Abstract:Purpose: Medical foundation models (FMs) offer a path to build high-performance diagnostic systems. However, their application to prostate cancer (PCa) detection from micro-ultrasound ({\mu}US) remains untested in clinical settings. We present ProstNFound+, an adaptation of FMs for PCa detection from {\mu}US, along with its first prospective validation. Methods: ProstNFound+ incorporates a medical FM, adapter tuning, and a custom prompt encoder that embeds PCa-specific clinical biomarkers. The model generates a cancer heatmap and a risk score for clinically significant PCa. Following training on multi-center retrospective data, the model is prospectively evaluated on data acquired five years later from a new clinical site. Model predictions are benchmarked against standard clinical scoring protocols (PRI-MUS and PI-RADS). Results: ProstNFound+ shows strong generalization to the prospective data, with no performance degradation compared to retrospective evaluation. It aligns closely with clinical scores and produces interpretable heatmaps consistent with biopsy-confirmed lesions. Conclusion: The results highlight its potential for clinical deployment, offering a scalable and interpretable alternative to expert-driven protocols.
TRUSWorthy: Toward Clinically Applicable Deep Learning for Confident Detection of Prostate Cancer in Micro-Ultrasound
Feb 20, 2025Abstract:While deep learning methods have shown great promise in improving the effectiveness of prostate cancer (PCa) diagnosis by detecting suspicious lesions from trans-rectal ultrasound (TRUS), they must overcome multiple simultaneous challenges. There is high heterogeneity in tissue appearance, significant class imbalance in favor of benign examples, and scarcity in the number and quality of ground truth annotations available to train models. Failure to address even a single one of these problems can result in unacceptable clinical outcomes.We propose TRUSWorthy, a carefully designed, tuned, and integrated system for reliable PCa detection. Our pipeline integrates self-supervised learning, multiple-instance learning aggregation using transformers, random-undersampled boosting and ensembling: these address label scarcity, weak labels, class imbalance, and overconfidence, respectively. We train and rigorously evaluate our method using a large, multi-center dataset of micro-ultrasound data. Our method outperforms previous state-of-the-art deep learning methods in terms of accuracy and uncertainty calibration, with AUROC and balanced accuracy scores of 79.9% and 71.5%, respectively. On the top 20% of predictions with the highest confidence, we can achieve a balanced accuracy of up to 91%. The success of TRUSWorthy demonstrates the potential of integrated deep learning solutions to meet clinical needs in a highly challenging deployment setting, and is a significant step towards creating a trustworthy system for computer-assisted PCa diagnosis.
Calibrated Diverse Ensemble Entropy Minimization for Robust Test-Time Adaptation in Prostate Cancer Detection
Jul 17, 2024



Abstract:High resolution micro-ultrasound has demonstrated promise in real-time prostate cancer detection, with deep learning becoming a prominent tool for learning complex tissue properties reflected on ultrasound. However, a significant roadblock to real-world deployment remains, which prior works often overlook: model performance suffers when applied to data from different clinical centers due to variations in data distribution. This distribution shift significantly impacts the model's robustness, posing major challenge to clinical deployment. Domain adaptation and specifically its test-time adaption (TTA) variant offer a promising solution to address this challenge. In a setting designed to reflect real-world conditions, we compare existing methods to state-of-the-art TTA approaches adopted for cancer detection, demonstrating the lack of robustness to distribution shifts in the former. We then propose Diverse Ensemble Entropy Minimization (DEnEM), questioning the effectiveness of current TTA methods on ultrasound data. We show that these methods, although outperforming baselines, are suboptimal due to relying on neural networks output probabilities, which could be uncalibrated, or relying on data augmentation, which is not straightforward to define on ultrasound data. Our results show a significant improvement of $5\%$ to $7\%$ in AUROC over the existing methods and $3\%$ to $5\%$ over TTA methods, demonstrating the advantage of DEnEM in addressing distribution shift. \keywords{Ultrasound Imaging \and Prostate Cancer \and Computer-aided Diagnosis \and Distribution Shift Robustness \and Test-time Adaptation.}
Benchmarking Image Transformers for Prostate Cancer Detection from Ultrasound Data
Mar 27, 2024Abstract:PURPOSE: Deep learning methods for classifying prostate cancer (PCa) in ultrasound images typically employ convolutional networks (CNNs) to detect cancer in small regions of interest (ROI) along a needle trace region. However, this approach suffers from weak labelling, since the ground-truth histopathology labels do not describe the properties of individual ROIs. Recently, multi-scale approaches have sought to mitigate this issue by combining the context awareness of transformers with a CNN feature extractor to detect cancer from multiple ROIs using multiple-instance learning (MIL). In this work, we present a detailed study of several image transformer architectures for both ROI-scale and multi-scale classification, and a comparison of the performance of CNNs and transformers for ultrasound-based prostate cancer classification. We also design a novel multi-objective learning strategy that combines both ROI and core predictions to further mitigate label noise. METHODS: We evaluate 3 image transformers on ROI-scale cancer classification, then use the strongest model to tune a multi-scale classifier with MIL. We train our MIL models using our novel multi-objective learning strategy and compare our results to existing baselines. RESULTS: We find that for both ROI-scale and multi-scale PCa detection, image transformer backbones lag behind their CNN counterparts. This deficit in performance is even more noticeable for larger models. When using multi-objective learning, we can improve performance of MIL, with a 77.9% AUROC, a sensitivity of 75.9%, and a specificity of 66.3%. CONCLUSION: Convolutional networks are better suited for modelling sparse datasets of prostate ultrasounds, producing more robust features than transformers in PCa detection. Multi-scale methods remain the best architecture for this task, with multi-objective learning presenting an effective way to improve performance.
TRUSformer: Improving Prostate Cancer Detection from Micro-Ultrasound Using Attention and Self-Supervision
Mar 03, 2023Abstract:A large body of previous machine learning methods for ultrasound-based prostate cancer detection classify small regions of interest (ROIs) of ultrasound signals that lie within a larger needle trace corresponding to a prostate tissue biopsy (called biopsy core). These ROI-scale models suffer from weak labeling as histopathology results available for biopsy cores only approximate the distribution of cancer in the ROIs. ROI-scale models do not take advantage of contextual information that are normally considered by pathologists, i.e. they do not consider information about surrounding tissue and larger-scale trends when identifying cancer. We aim to improve cancer detection by taking a multi-scale, i.e. ROI-scale and biopsy core-scale, approach. Methods: Our multi-scale approach combines (i) an "ROI-scale" model trained using self-supervised learning to extract features from small ROIs and (ii) a "core-scale" transformer model that processes a collection of extracted features from multiple ROIs in the needle trace region to predict the tissue type of the corresponding core. Attention maps, as a byproduct, allow us to localize cancer at the ROI scale. We analyze this method using a dataset of micro-ultrasound acquired from 578 patients who underwent prostate biopsy, and compare our model to baseline models and other large-scale studies in the literature. Results and Conclusions: Our model shows consistent and substantial performance improvements compared to ROI-scale-only models. It achieves 80.3% AUROC, a statistically significant improvement over ROI-scale classification. We also compare our method to large studies on prostate cancer detection, using other imaging modalities. Our code is publicly available at www.github.com/med-i-lab/TRUSFormer
Self-Supervised Learning with Limited Labeled Data for Prostate Cancer Detection in High Frequency Ultrasound
Nov 01, 2022Abstract:Deep learning-based analysis of high-frequency, high-resolution micro-ultrasound data shows great promise for prostate cancer detection. Previous approaches to analysis of ultrasound data largely follow a supervised learning paradigm. Ground truth labels for ultrasound images used for training deep networks often include coarse annotations generated from the histopathological analysis of tissue samples obtained via biopsy. This creates inherent limitations on the availability and quality of labeled data, posing major challenges to the success of supervised learning methods. On the other hand, unlabeled prostate ultrasound data are more abundant. In this work, we successfully apply self-supervised representation learning to micro-ultrasound data. Using ultrasound data from 1028 biopsy cores of 391 subjects obtained in two clinical centres, we demonstrate that feature representations learnt with this method can be used to classify cancer from non-cancer tissue, obtaining an AUROC score of 91% on an independent test set. To the best of our knowledge, this is the first successful end-to-end self-supervised learning approach for prostate cancer detection using ultrasound data. Our method outperforms baseline supervised learning approaches, generalizes well between different data centers, and scale well in performance as more unlabeled data are added, making it a promising approach for future research using large volumes of unlabeled data.
Towards Confident Detection of Prostate Cancer using High Resolution Micro-ultrasound
Jul 21, 2022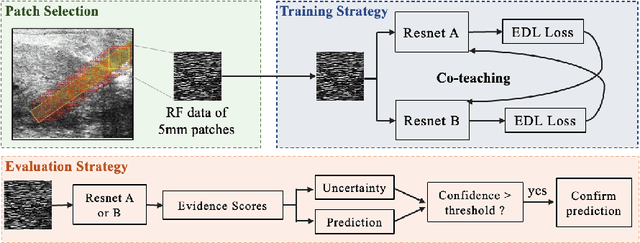
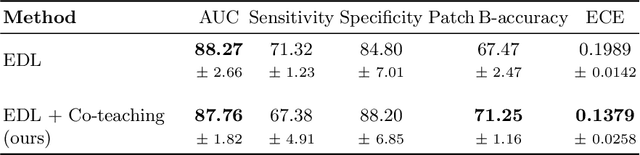
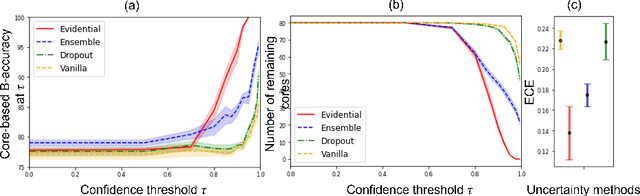
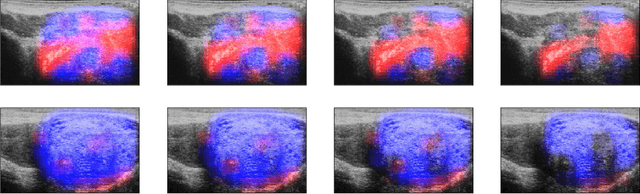
Abstract:MOTIVATION: Detection of prostate cancer during transrectal ultrasound-guided biopsy is challenging. The highly heterogeneous appearance of cancer, presence of ultrasound artefacts, and noise all contribute to these difficulties. Recent advancements in high-frequency ultrasound imaging - micro-ultrasound - have drastically increased the capability of tissue imaging at high resolution. Our aim is to investigate the development of a robust deep learning model specifically for micro-ultrasound-guided prostate cancer biopsy. For the model to be clinically adopted, a key challenge is to design a solution that can confidently identify the cancer, while learning from coarse histopathology measurements of biopsy samples that introduce weak labels. METHODS: We use a dataset of micro-ultrasound images acquired from 194 patients, who underwent prostate biopsy. We train a deep model using a co-teaching paradigm to handle noise in labels, together with an evidential deep learning method for uncertainty estimation. We evaluate the performance of our model using the clinically relevant metric of accuracy vs. confidence. RESULTS: Our model achieves a well-calibrated estimation of predictive uncertainty with area under the curve of 88$\%$. The use of co-teaching and evidential deep learning in combination yields significantly better uncertainty estimation than either alone. We also provide a detailed comparison against state-of-the-art in uncertainty estimation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge