Matthew Lee
Learning to Represent Individual Differences for Choice Decision Making
Mar 27, 2025Abstract:Human decision making can be challenging to predict because decisions are affected by a number of complex factors. Adding to this complexity, decision-making processes can differ considerably between individuals, and methods aimed at predicting human decisions need to take individual differences into account. Behavioral science offers methods by which to measure individual differences (e.g., questionnaires, behavioral models), but these are often narrowed down to low dimensions and not tailored to specific prediction tasks. This paper investigates the use of representation learning to measure individual differences from behavioral experiment data. Representation learning offers a flexible approach to create individual embeddings from data that are both structured (e.g., demographic information) and unstructured (e.g., free text), where the flexibility provides more options for individual difference measures for personalization, e.g., free text responses may allow for open-ended questions that are less privacy-sensitive. In the current paper we use representation learning to characterize individual differences in human performance on an economic decision-making task. We demonstrate that models using representation learning to capture individual differences consistently improve decision predictions over models without representation learning, and even outperform well-known theory-based behavioral models used in these environments. Our results propose that representation learning offers a useful and flexible tool to capture individual differences.
MusicFlow: Cascaded Flow Matching for Text Guided Music Generation
Oct 27, 2024



Abstract:We introduce MusicFlow, a cascaded text-to-music generation model based on flow matching. Based on self-supervised representations to bridge between text descriptions and music audios, we construct two flow matching networks to model the conditional distribution of semantic and acoustic features. Additionally, we leverage masked prediction as the training objective, enabling the model to generalize to other tasks such as music infilling and continuation in a zero-shot manner. Experiments on MusicCaps reveal that the music generated by MusicFlow exhibits superior quality and text coherence despite being over $2\sim5$ times smaller and requiring $5$ times fewer iterative steps. Simultaneously, the model can perform other music generation tasks and achieves competitive performance in music infilling and continuation. Our code and model will be publicly available.
Training Towards Critical Use: Learning to Situate AI Predictions Relative to Human Knowledge
Aug 30, 2023
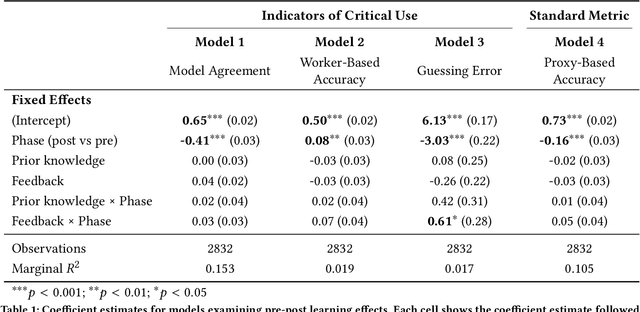
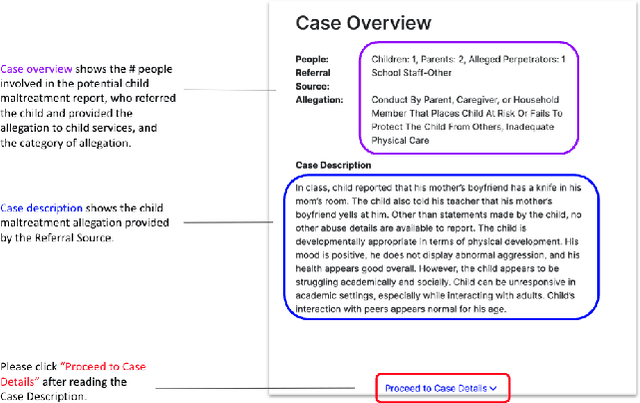
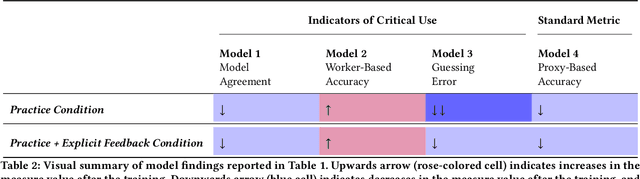
Abstract:A growing body of research has explored how to support humans in making better use of AI-based decision support, including via training and onboarding. Existing research has focused on decision-making tasks where it is possible to evaluate "appropriate reliance" by comparing each decision against a ground truth label that cleanly maps to both the AI's predictive target and the human decision-maker's goals. However, this assumption does not hold in many real-world settings where AI tools are deployed today (e.g., social work, criminal justice, and healthcare). In this paper, we introduce a process-oriented notion of appropriate reliance called critical use that centers the human's ability to situate AI predictions against knowledge that is uniquely available to them but unavailable to the AI model. To explore how training can support critical use, we conduct a randomized online experiment in a complex social decision-making setting: child maltreatment screening. We find that, by providing participants with accelerated, low-stakes opportunities to practice AI-assisted decision-making in this setting, novices came to exhibit patterns of disagreement with AI that resemble those of experienced workers. A qualitative examination of participants' explanations for their AI-assisted decisions revealed that they drew upon qualitative case narratives, to which the AI model did not have access, to learn when (not) to rely on AI predictions. Our findings open new questions for the study and design of training for real-world AI-assisted decision-making.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Probabilistic 3D surface reconstruction from sparse MRI information
Oct 05, 2020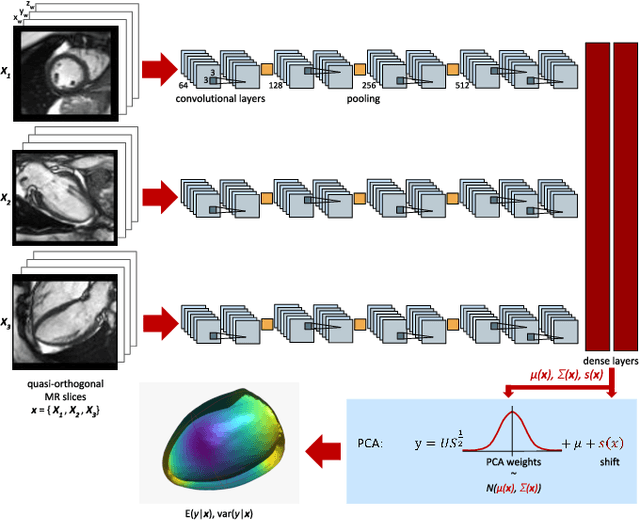
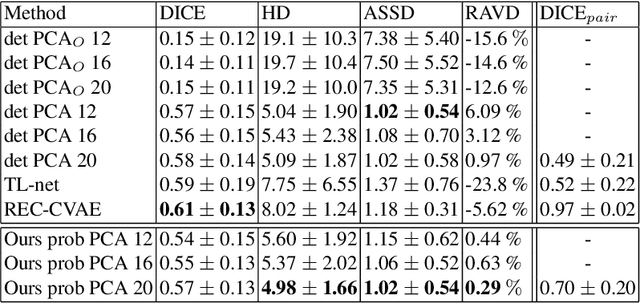

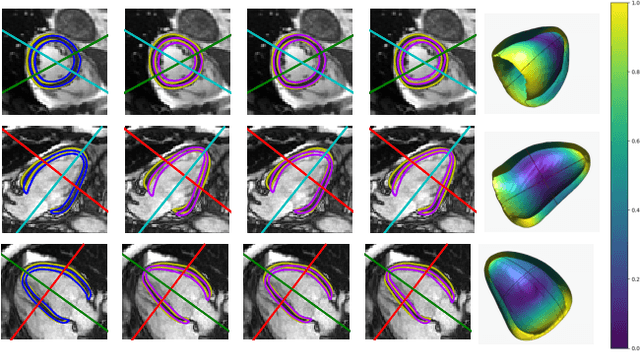
Abstract:Surface reconstruction from magnetic resonance (MR) imaging data is indispensable in medical image analysis and clinical research. A reliable and effective reconstruction tool should: be fast in prediction of accurate well localised and high resolution models, evaluate prediction uncertainty, work with as little input data as possible. Current deep learning state of the art (SOTA) 3D reconstruction methods, however, often only produce shapes of limited variability positioned in a canonical position or lack uncertainty evaluation. In this paper, we present a novel probabilistic deep learning approach for concurrent 3D surface reconstruction from sparse 2D MR image data and aleatoric uncertainty prediction. Our method is capable of reconstructing large surface meshes from three quasi-orthogonal MR imaging slices from limited training sets whilst modelling the location of each mesh vertex through a Gaussian distribution. Prior shape information is encoded using a built-in linear principal component analysis (PCA) model. Extensive experiments on cardiac MR data show that our probabilistic approach successfully assesses prediction uncertainty while at the same time qualitatively and quantitatively outperforms SOTA methods in shape prediction. Compared to SOTA, we are capable of properly localising and orientating the prediction via the use of a spatially aware neural network.
One-Shot Learning for Language Modelling
Jul 19, 2020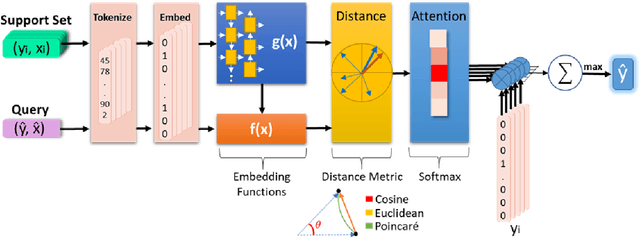

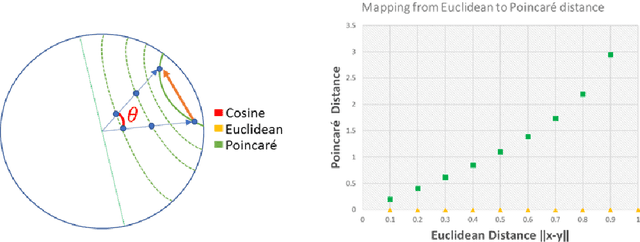
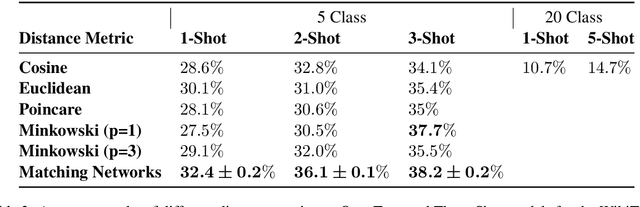
Abstract:Humans can infer a great deal about the meaning of a word, using the syntax and semantics of surrounding words even if it is their first time reading or hearing it. We can also generalise the learned concept of the word to new tasks. Despite great progress in achieving human-level performance in certain tasks (Silver et al., 2016), learning from one or few examples remains a key challenge in machine learning, and has not thoroughly been explored in Natural Language Processing (NLP). In this work we tackle the problem of oneshot learning for an NLP task by employing ideas from recent developments in machine learning: embeddings, attention mechanisms (softmax) and similarity measures (cosine, Euclidean, Poincare, and Minkowski). We adapt the framework suggested in matching networks (Vinyals et al., 2016), and explore the effectiveness of the aforementioned methods in one, two and three-shot learning problems on the task of predicting missing word explored in (Vinyals et al., 2016) by using the WikiText-2 dataset. Our work contributes in two ways: Our first contribution is that we explore the effectiveness of different distance metrics on k-shot learning, and show that there is no single best distance metric for k-shot learning, which challenges common belief. We found that the performance of a distance metric depends on the number of shots used during training. The second contribution of our work is that we establish a benchmark for one, two, and three-shot learning on a language task with a publicly available dataset that can be used to benchmark against in future research.
Detecting Affective Flow States of Knowledge Workers Using Physiological Sensors
Jun 18, 2020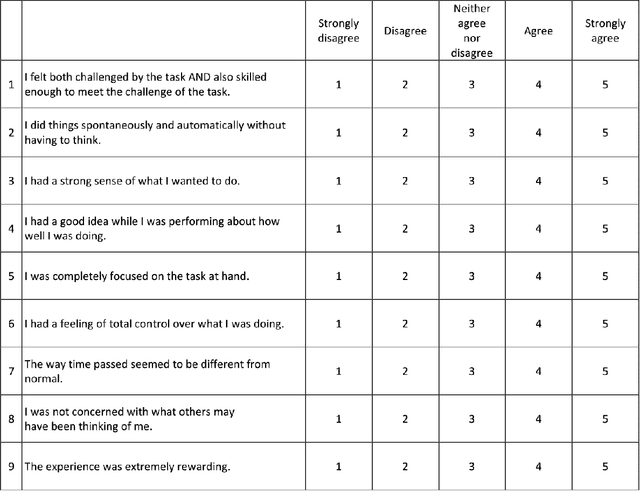
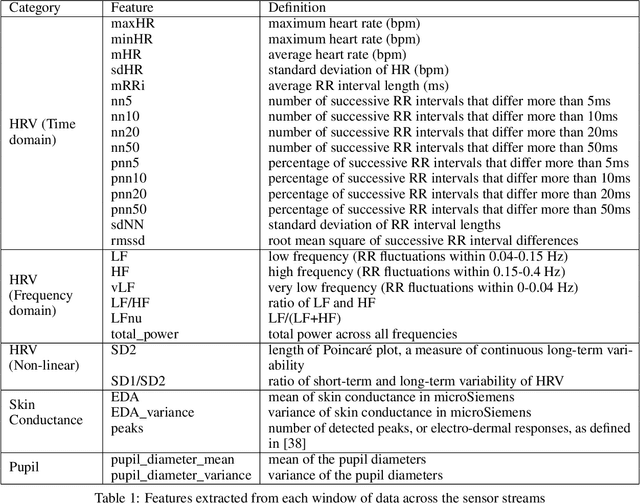


Abstract:Flow-like experiences at work are important for productivity and worker well-being. However, it is difficult to objectively detect when workers are experiencing flow in their work. In this paper, we investigate how to predict a worker's focus state based on physiological signals. We conducted a lab study to collect physiological data from knowledge workers experienced different levels of flow while performing work tasks. We used the nine characteristics of flow to design tasks that would induce different focus states. A manipulation check using the Flow Short Scale verified that participants experienced three distinct flow states, one overly challenging non-flow state, and two types of flow states, balanced flow, and automatic flow. We built machine learning classifiers that can distinguish between non-flow and flow states with 0.889 average AUC and rest states from working states with 0.98 average AUC. The results show that physiological sensing can detect focused flow states of knowledge workers and can enable ways to for individuals and organizations to improve both productivity and worker satisfaction.
Pain Intensity Estimation from Mobile Video Using 2D and 3D Facial Keypoints
Jun 17, 2020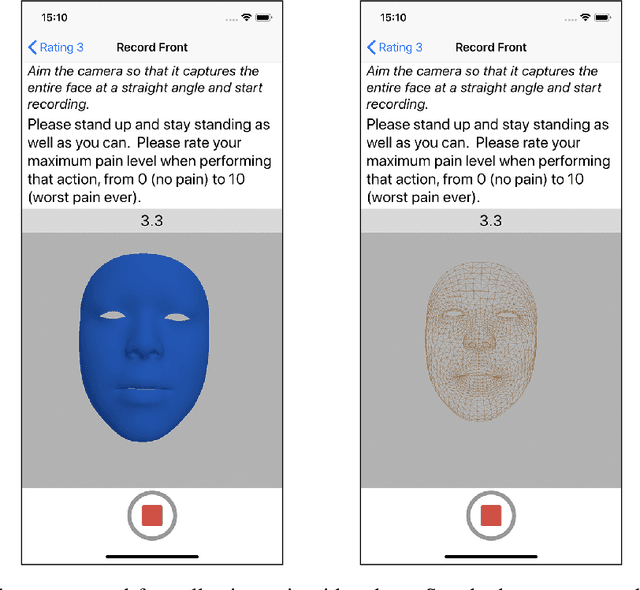
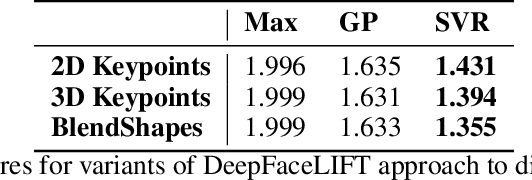
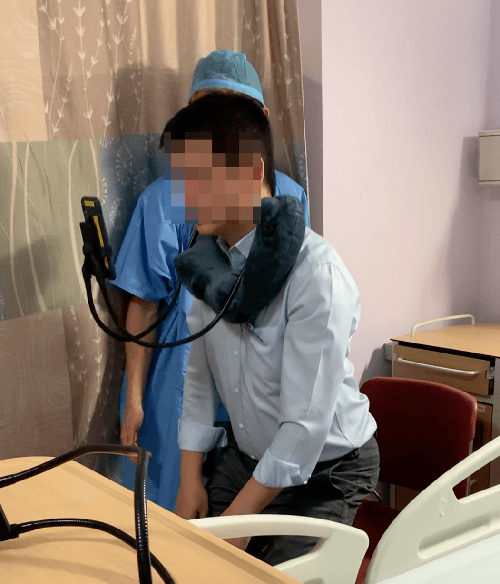
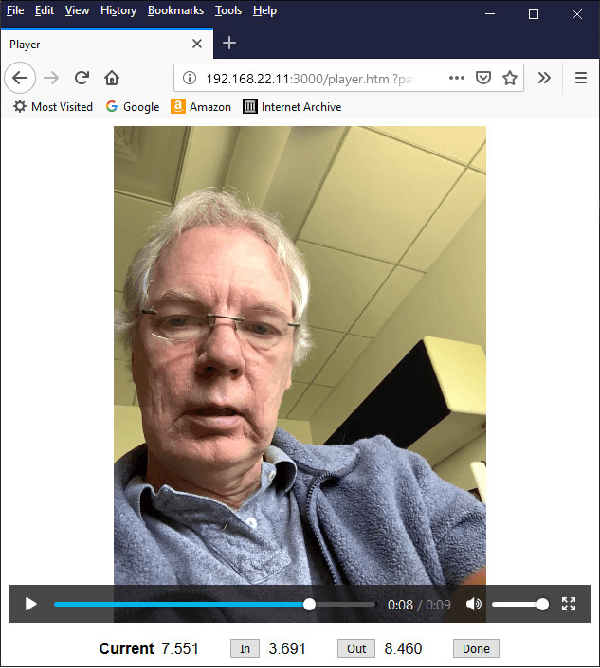
Abstract:Managing post-surgical pain is critical for successful surgical outcomes. One of the challenges of pain management is accurately assessing the pain level of patients. Self-reported numeric pain ratings are limited because they are subjective, can be affected by mood, and can influence the patient's perception of pain when making comparisons. In this paper, we introduce an approach that analyzes 2D and 3D facial keypoints of post-surgical patients to estimate their pain intensity level. Our approach leverages the previously unexplored capabilities of a smartphone to capture a dense 3D representation of a person's face as input for pain intensity level estimation. Our contributions are adata collection study with post-surgical patients to collect ground-truth labeled sequences of 2D and 3D facial keypoints for developing a pain estimation algorithm, a pain estimation model that uses multiple instance learning to overcome inherent limitations in facial keypoint sequences, and the preliminary results of the pain estimation model using 2D and 3D features with comparisons of alternate approaches.
A Summary of the First Workshop on Language Technology for Language Documentation and Revitalization
Apr 27, 2020

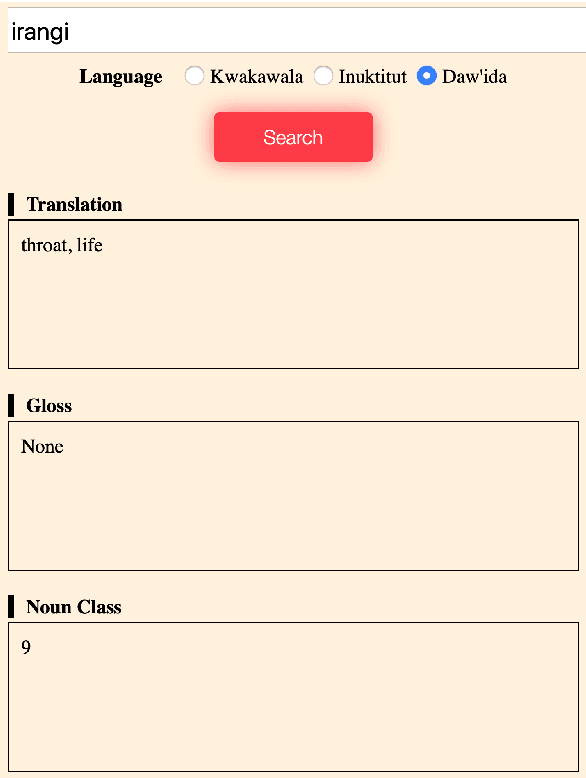
Abstract:Despite recent advances in natural language processing and other language technology, the application of such technology to language documentation and conservation has been limited. In August 2019, a workshop was held at Carnegie Mellon University in Pittsburgh to attempt to bring together language community members, documentary linguists, and technologists to discuss how to bridge this gap and create prototypes of novel and practical language revitalization technologies. This paper reports the results of this workshop, including issues discussed, and various conceived and implemented technologies for nine languages: Arapaho, Cayuga, Inuktitut, Irish Gaelic, Kidaw'ida, Kwak'wala, Ojibwe, San Juan Quiahije Chatino, and Seneca.
Universal Phone Recognition with a Multilingual Allophone System
Feb 26, 2020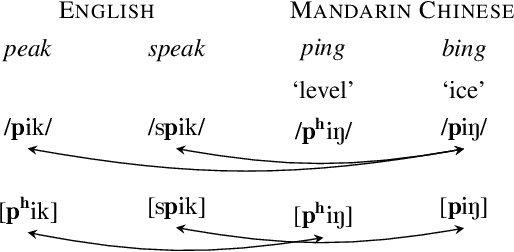
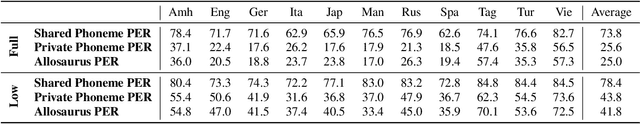
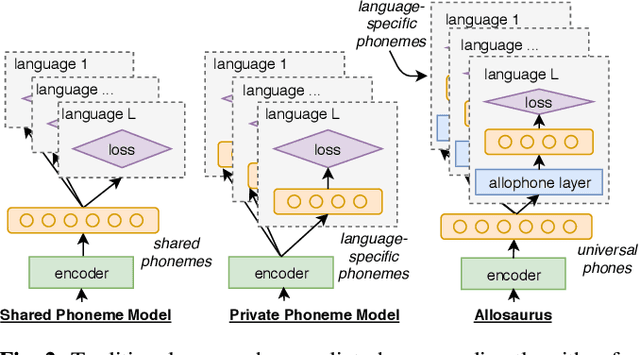
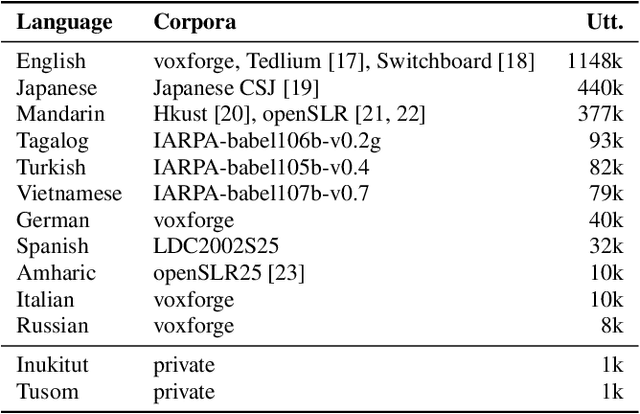
Abstract:Multilingual models can improve language processing, particularly for low resource situations, by sharing parameters across languages. Multilingual acoustic models, however, generally ignore the difference between phonemes (sounds that can support lexical contrasts in a particular language) and their corresponding phones (the sounds that are actually spoken, which are language independent). This can lead to performance degradation when combining a variety of training languages, as identically annotated phonemes can actually correspond to several different underlying phonetic realizations. In this work, we propose a joint model of both language-independent phone and language-dependent phoneme distributions. In multilingual ASR experiments over 11 languages, we find that this model improves testing performance by 2% phoneme error rate absolute in low-resource conditions. Additionally, because we are explicitly modeling language-independent phones, we can build a (nearly-)universal phone recognizer that, when combined with the PHOIBLE large, manually curated database of phone inventories, can be customized into 2,000 language dependent recognizers. Experiments on two low-resourced indigenous languages, Inuktitut and Tusom, show that our recognizer achieves phone accuracy improvements of more than 17%, moving a step closer to speech recognition for all languages in the world.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge