Ziyang Xu
ERNIE 5.0 Technical Report
Feb 04, 2026Abstract:In this report, we introduce ERNIE 5.0, a natively autoregressive foundation model desinged for unified multimodal understanding and generation across text, image, video, and audio. All modalities are trained from scratch under a unified next-group-of-tokens prediction objective, based on an ultra-sparse mixture-of-experts (MoE) architecture with modality-agnostic expert routing. To address practical challenges in large-scale deployment under diverse resource constraints, ERNIE 5.0 adopts a novel elastic training paradigm. Within a single pre-training run, the model learns a family of sub-models with varying depths, expert capacities, and routing sparsity, enabling flexible trade-offs among performance, model size, and inference latency in memory- or time-constrained scenarios. Moreover, we systematically address the challenges of scaling reinforcement learning to unified foundation models, thereby guaranteeing efficient and stable post-training under ultra-sparse MoE architectures and diverse multimodal settings. Extensive experiments demonstrate that ERNIE 5.0 achieves strong and balanced performance across multiple modalities. To the best of our knowledge, among publicly disclosed models, ERNIE 5.0 represents the first production-scale realization of a trillion-parameter unified autoregressive model that supports both multimodal understanding and generation. To facilitate further research, we present detailed visualizations of modality-agnostic expert routing in the unified model, alongside comprehensive empirical analysis of elastic training, aiming to offer profound insights to the community.
Establishing dermatopathology encyclopedia DermpathNet with Artificial Intelligence-Based Workflow
Jan 27, 2026Abstract:Accessing high-quality, open-access dermatopathology image datasets for learning and cross-referencing is a common challenge for clinicians and dermatopathology trainees. To establish a comprehensive open-access dermatopathology dataset for educational, cross-referencing, and machine-learning purposes, we employed a hybrid workflow to curate and categorize images from the PubMed Central (PMC) repository. We used specific keywords to extract relevant images, and classified them using a novel hybrid method that combined deep learning-based image modality classification with figure caption analyses. Validation on 651 manually annotated images demonstrated the robustness of our workflow, with an F-score of 89.6\% for the deep learning approach, 61.0\% for the keyword-based retrieval method, and 90.4\% for the hybrid approach. We retrieved over 7,772 images across 166 diagnoses and released this fully annotated dataset, reviewed by board-certified dermatopathologists. Using our dataset as a challenging task, we found the current image analysis algorithm from OpenAI inadequate for analyzing dermatopathology images. In conclusion, we have developed a large, peer-reviewed, open-access dermatopathology image dataset, DermpathNet, which features a semi-automated curation workflow.
All That Glisters Is Not Gold: A Benchmark for Reference-Free Counterfactual Financial Misinformation Detection
Jan 08, 2026Abstract:We introduce RFC Bench, a benchmark for evaluating large language models on financial misinformation under realistic news. RFC Bench operates at the paragraph level and captures the contextual complexity of financial news where meaning emerges from dispersed cues. The benchmark defines two complementary tasks: reference free misinformation detection and comparison based diagnosis using paired original perturbed inputs. Experiments reveal a consistent pattern: performance is substantially stronger when comparative context is available, while reference free settings expose significant weaknesses, including unstable predictions and elevated invalid outputs. These results indicate that current models struggle to maintain coherent belief states without external grounding. By highlighting this gap, RFC Bench provides a structured testbed for studying reference free reasoning and advancing more reliable financial misinformation detection in real world settings.
Genesis: Multimodal Driving Scene Generation with Spatio-Temporal and Cross-Modal Consistency
Jun 09, 2025Abstract:We present Genesis, a unified framework for joint generation of multi-view driving videos and LiDAR sequences with spatio-temporal and cross-modal consistency. Genesis employs a two-stage architecture that integrates a DiT-based video diffusion model with 3D-VAE encoding, and a BEV-aware LiDAR generator with NeRF-based rendering and adaptive sampling. Both modalities are directly coupled through a shared latent space, enabling coherent evolution across visual and geometric domains. To guide the generation with structured semantics, we introduce DataCrafter, a captioning module built on vision-language models that provides scene-level and instance-level supervision. Extensive experiments on the nuScenes benchmark demonstrate that Genesis achieves state-of-the-art performance across video and LiDAR metrics (FVD 16.95, FID 4.24, Chamfer 0.611), and benefits downstream tasks including segmentation and 3D detection, validating the semantic fidelity and practical utility of the generated data.
REACT: Representation Extraction And Controllable Tuning to Overcome Overfitting in LLM Knowledge Editing
May 25, 2025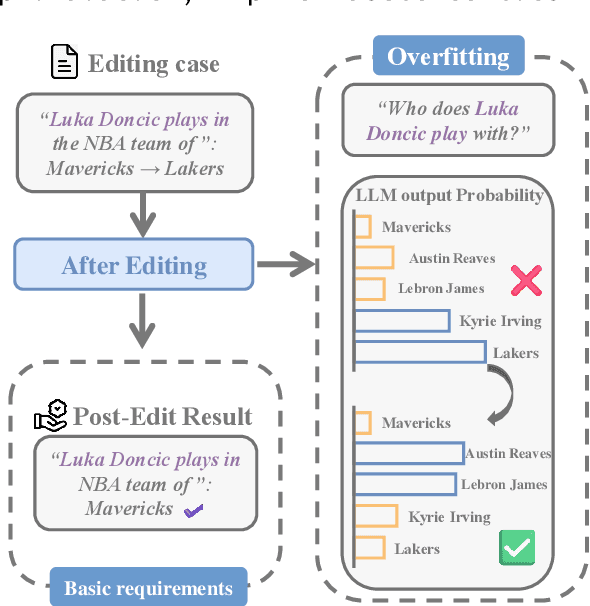
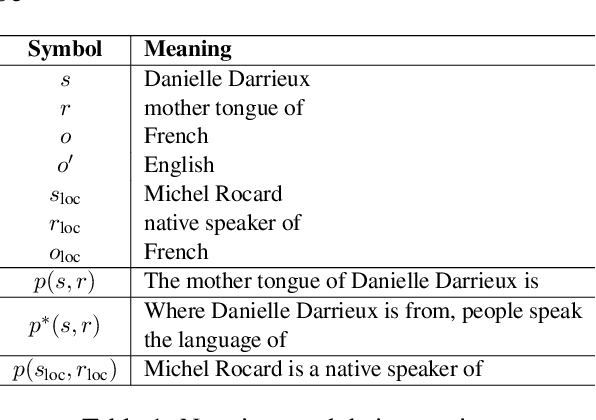
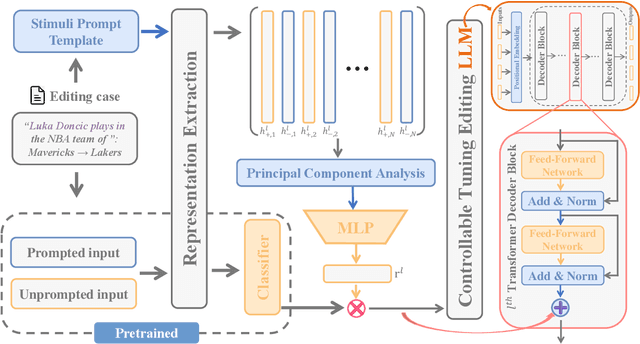
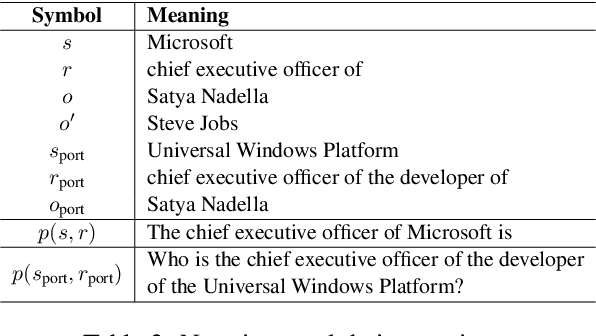
Abstract:Large language model editing methods frequently suffer from overfitting, wherein factual updates can propagate beyond their intended scope, overemphasizing the edited target even when it's contextually inappropriate. To address this challenge, we introduce REACT (Representation Extraction And Controllable Tuning), a unified two-phase framework designed for precise and controllable knowledge editing. In the initial phase, we utilize tailored stimuli to extract latent factual representations and apply Principal Component Analysis with a simple learnbale linear transformation to compute a directional "belief shift" vector for each instance. In the second phase, we apply controllable perturbations to hidden states using the obtained vector with a magnitude scalar, gated by a pre-trained classifier that permits edits only when contextually necessary. Relevant experiments on EVOKE benchmarks demonstrate that REACT significantly reduces overfitting across nearly all evaluation metrics, and experiments on COUNTERFACT and MQuAKE shows that our method preserves balanced basic editing performance (reliability, locality, and generality) under diverse editing scenarios.
TUM2TWIN: Introducing the Large-Scale Multimodal Urban Digital Twin Benchmark Dataset
May 13, 2025



Abstract:Urban Digital Twins (UDTs) have become essential for managing cities and integrating complex, heterogeneous data from diverse sources. Creating UDTs involves challenges at multiple process stages, including acquiring accurate 3D source data, reconstructing high-fidelity 3D models, maintaining models' updates, and ensuring seamless interoperability to downstream tasks. Current datasets are usually limited to one part of the processing chain, hampering comprehensive UDTs validation. To address these challenges, we introduce the first comprehensive multimodal Urban Digital Twin benchmark dataset: TUM2TWIN. This dataset includes georeferenced, semantically aligned 3D models and networks along with various terrestrial, mobile, aerial, and satellite observations boasting 32 data subsets over roughly 100,000 $m^2$ and currently 767 GB of data. By ensuring georeferenced indoor-outdoor acquisition, high accuracy, and multimodal data integration, the benchmark supports robust analysis of sensors and the development of advanced reconstruction methods. Additionally, we explore downstream tasks demonstrating the potential of TUM2TWIN, including novel view synthesis of NeRF and Gaussian Splatting, solar potential analysis, point cloud semantic segmentation, and LoD3 building reconstruction. We are convinced this contribution lays a foundation for overcoming current limitations in UDT creation, fostering new research directions and practical solutions for smarter, data-driven urban environments. The project is available under: https://tum2t.win
PixelHacker: Image Inpainting with Structural and Semantic Consistency
Apr 30, 2025Abstract:Image inpainting is a fundamental research area between image editing and image generation. Recent state-of-the-art (SOTA) methods have explored novel attention mechanisms, lightweight architectures, and context-aware modeling, demonstrating impressive performance. However, they often struggle with complex structure (e.g., texture, shape, spatial relations) and semantics (e.g., color consistency, object restoration, and logical correctness), leading to artifacts and inappropriate generation. To address this challenge, we design a simple yet effective inpainting paradigm called latent categories guidance, and further propose a diffusion-based model named PixelHacker. Specifically, we first construct a large dataset containing 14 million image-mask pairs by annotating foreground and background (potential 116 and 21 categories, respectively). Then, we encode potential foreground and background representations separately through two fixed-size embeddings, and intermittently inject these features into the denoising process via linear attention. Finally, by pre-training on our dataset and fine-tuning on open-source benchmarks, we obtain PixelHacker. Extensive experiments show that PixelHacker comprehensively outperforms the SOTA on a wide range of datasets (Places2, CelebA-HQ, and FFHQ) and exhibits remarkable consistency in both structure and semantics. Project page at https://hustvl.github.io/PixelHacker.
MedReason: Eliciting Factual Medical Reasoning Steps in LLMs via Knowledge Graphs
Apr 01, 2025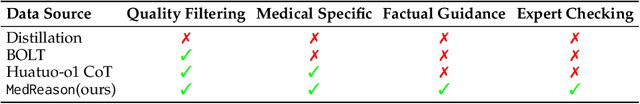
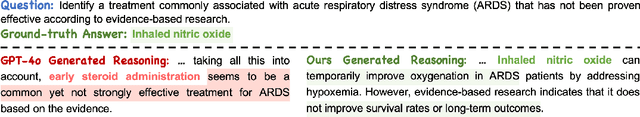
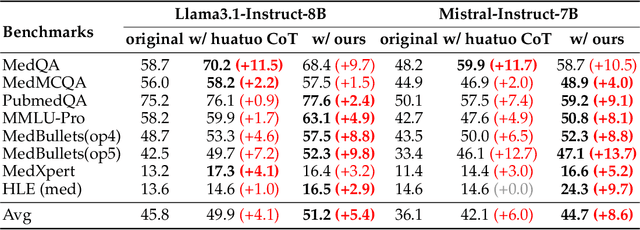
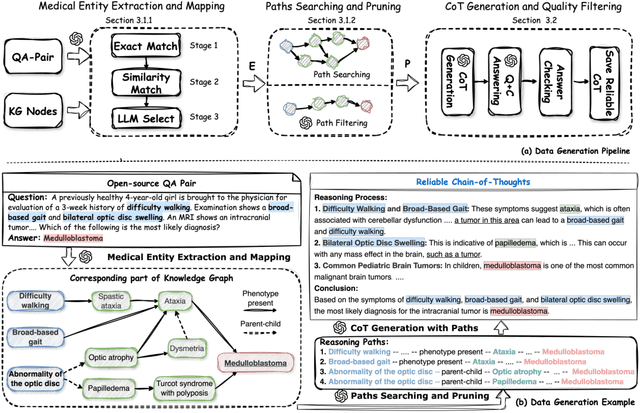
Abstract:Medical tasks such as diagnosis and treatment planning require precise and complex reasoning, particularly in life-critical domains. Unlike mathematical reasoning, medical reasoning demands meticulous, verifiable thought processes to ensure reliability and accuracy. However, there is a notable lack of datasets that provide transparent, step-by-step reasoning to validate and enhance the medical reasoning ability of AI models. To bridge this gap, we introduce MedReason, a large-scale high-quality medical reasoning dataset designed to enable faithful and explainable medical problem-solving in large language models (LLMs). We utilize a structured medical knowledge graph (KG) to convert clinical QA pairs into logical chains of reasoning, or ``thinking paths'', which trace connections from question elements to answers via relevant KG entities. Each path is validated for consistency with clinical logic and evidence-based medicine. Our pipeline generates detailed reasoning for various medical questions from 7 medical datasets, resulting in a dataset of 32,682 question-answer pairs, each with detailed, step-by-step explanations. Experiments demonstrate that fine-tuning with our dataset consistently boosts medical problem-solving capabilities, achieving significant gains of up to 7.7% for DeepSeek-Ditill-8B. Our top-performing model, MedReason-8B, outperforms the Huatuo-o1-8B, a state-of-the-art medical reasoning model, by up to 4.2% on the clinical benchmark MedBullets. We also engage medical professionals from diverse specialties to assess our dataset's quality, ensuring MedReason offers accurate and coherent medical reasoning. Our data, models, and code will be publicly available.
PDeepPP:A Deep learning framework with Pretrained Protein language for peptide classification
Feb 21, 2025Abstract:Protein post-translational modifications (PTMs) and bioactive peptides (BPs) play critical roles in various biological processes and have significant therapeutic potential. However, identifying PTM sites and bioactive peptides through experimental methods is often labor-intensive, costly, and time-consuming. As a result, computational tools, particularly those based on deep learning, have become effective solutions for predicting PTM sites and peptide bioactivity. Despite progress in this field, existing methods still struggle with the complexity of protein sequences and the challenge of requiring high-quality predictions across diverse datasets. To address these issues, we propose a deep learning framework that integrates pretrained protein language models with a neural network combining transformer and CNN for peptide classification. By leveraging the ability of pretrained models to capture complex relationships within protein sequences, combined with the predictive power of parallel networks, our approach improves feature extraction while enhancing prediction accuracy. This framework was applied to multiple tasks involving PTM site and bioactive peptide prediction, utilizing large-scale datasets to enhance the model's robustness. In the comparison across 33 tasks, the model achieved state-of-the-art (SOTA) performance in 25 of them, surpassing existing methods and demonstrating its versatility across different datasets. Our results suggest that this approach provides a scalable and effective solution for large-scale peptide discovery and PTM analysis, paving the way for more efficient peptide classification and functional annotation.
A foundation model for human-AI collaboration in medical literature mining
Jan 27, 2025



Abstract:Systematic literature review is essential for evidence-based medicine, requiring comprehensive analysis of clinical trial publications. However, the application of artificial intelligence (AI) models for medical literature mining has been limited by insufficient training and evaluation across broad therapeutic areas and diverse tasks. Here, we present LEADS, an AI foundation model for study search, screening, and data extraction from medical literature. The model is trained on 633,759 instruction data points in LEADSInstruct, curated from 21,335 systematic reviews, 453,625 clinical trial publications, and 27,015 clinical trial registries. We showed that LEADS demonstrates consistent improvements over four cutting-edge generic large language models (LLMs) on six tasks. Furthermore, LEADS enhances expert workflows by providing supportive references following expert requests, streamlining processes while maintaining high-quality results. A study with 16 clinicians and medical researchers from 14 different institutions revealed that experts collaborating with LEADS achieved a recall of 0.81 compared to 0.77 experts working alone in study selection, with a time savings of 22.6%. In data extraction tasks, experts using LEADS achieved an accuracy of 0.85 versus 0.80 without using LEADS, alongside a 26.9% time savings. These findings highlight the potential of specialized medical literature foundation models to outperform generic models, delivering significant quality and efficiency benefits when integrated into expert workflows for medical literature mining.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge