Yuchen Ren
Probing Scientific General Intelligence of LLMs with Scientist-Aligned Workflows
Dec 18, 2025Abstract:Despite advances in scientific AI, a coherent framework for Scientific General Intelligence (SGI)-the ability to autonomously conceive, investigate, and reason across scientific domains-remains lacking. We present an operational SGI definition grounded in the Practical Inquiry Model (PIM: Deliberation, Conception, Action, Perception) and operationalize it via four scientist-aligned tasks: deep research, idea generation, dry/wet experiments, and experimental reasoning. SGI-Bench comprises over 1,000 expert-curated, cross-disciplinary samples inspired by Science's 125 Big Questions, enabling systematic evaluation of state-of-the-art LLMs. Results reveal gaps: low exact match (10--20%) in deep research despite step-level alignment; ideas lacking feasibility and detail; high code executability but low execution result accuracy in dry experiments; low sequence fidelity in wet protocols; and persistent multimodal comparative-reasoning challenges. We further introduce Test-Time Reinforcement Learning (TTRL), which optimizes retrieval-augmented novelty rewards at inference, enhancing hypothesis novelty without reference answer. Together, our PIM-grounded definition, workflow-centric benchmark, and empirical insights establish a foundation for AI systems that genuinely participate in scientific discovery.
ChronusOmni: Improving Time Awareness of Omni Large Language Models
Dec 10, 2025Abstract:Time awareness is a fundamental ability of omni large language models, especially for understanding long videos and answering complex questions. Previous approaches mainly target vision-language scenarios and focus on the explicit temporal grounding questions, such as identifying when a visual event occurs or determining what event happens at aspecific time. However, they often make insufficient use of the audio modality, and overlook implicit temporal grounding across modalities--for example, identifying what is visually present when a character speaks, or determining what is said when a visual event occurs--despite such cross-modal temporal relations being prevalent in real-world scenarios. In this paper, we propose ChronusOmni, an omni large language model designed to enhance temporal awareness for both explicit and implicit audiovisual temporal grounding. First, we interleave text-based timestamp tokens with visual and audio representations at each time unit, enabling unified temporal modeling across modalities. Second, to enforce correct temporal ordering and strengthen fine-grained temporal reasoning, we incorporate reinforcement learning with specially designed reward functions. Moreover, we construct ChronusAV, a temporally-accurate, modality-complete, and cross-modal-aligned dataset to support the training and evaluation on audiovisual temporal grounding task. Experimental results demonstrate that ChronusOmni achieves state-of-the-art performance on ChronusAV with more than 30% improvement and top results on most metrics upon other temporal grounding benchmarks. This highlights the strong temporal awareness of our model across modalities, while preserving general video and audio understanding capabilities.
KORE: Enhancing Knowledge Injection for Large Multimodal Models via Knowledge-Oriented Augmentations and Constraints
Oct 22, 2025Abstract:Large Multimodal Models encode extensive factual knowledge in their pre-trained weights. However, its knowledge remains static and limited, unable to keep pace with real-world developments, which hinders continuous knowledge acquisition. Effective knowledge injection thus becomes critical, involving two goals: knowledge adaptation (injecting new knowledge) and knowledge retention (preserving old knowledge). Existing methods often struggle to learn new knowledge and suffer from catastrophic forgetting. To address this, we propose KORE, a synergistic method of KnOwledge-oRientEd augmentations and constraints for injecting new knowledge into large multimodal models while preserving old knowledge. Unlike general text or image data augmentation, KORE automatically converts individual knowledge items into structured and comprehensive knowledge to ensure that the model accurately learns new knowledge, enabling accurate adaptation. Meanwhile, KORE stores previous knowledge in the covariance matrix of LMM's linear layer activations and initializes the adapter by projecting the original weights into the matrix's null space, defining a fine-tuning direction that minimizes interference with previous knowledge, enabling powerful retention. Extensive experiments on various LMMs, including LLaVA-v1.5-7B, LLaVA-v1.5-13B, and Qwen2.5-VL-7B, show that KORE achieves superior new knowledge injection performance and effectively mitigates catastrophic forgetting.
A Survey of Scientific Large Language Models: From Data Foundations to Agent Frontiers
Aug 28, 2025



Abstract:Scientific Large Language Models (Sci-LLMs) are transforming how knowledge is represented, integrated, and applied in scientific research, yet their progress is shaped by the complex nature of scientific data. This survey presents a comprehensive, data-centric synthesis that reframes the development of Sci-LLMs as a co-evolution between models and their underlying data substrate. We formulate a unified taxonomy of scientific data and a hierarchical model of scientific knowledge, emphasizing the multimodal, cross-scale, and domain-specific challenges that differentiate scientific corpora from general natural language processing datasets. We systematically review recent Sci-LLMs, from general-purpose foundations to specialized models across diverse scientific disciplines, alongside an extensive analysis of over 270 pre-/post-training datasets, showing why Sci-LLMs pose distinct demands -- heterogeneous, multi-scale, uncertainty-laden corpora that require representations preserving domain invariance and enabling cross-modal reasoning. On evaluation, we examine over 190 benchmark datasets and trace a shift from static exams toward process- and discovery-oriented assessments with advanced evaluation protocols. These data-centric analyses highlight persistent issues in scientific data development and discuss emerging solutions involving semi-automated annotation pipelines and expert validation. Finally, we outline a paradigm shift toward closed-loop systems where autonomous agents based on Sci-LLMs actively experiment, validate, and contribute to a living, evolving knowledge base. Collectively, this work provides a roadmap for building trustworthy, continually evolving artificial intelligence (AI) systems that function as a true partner in accelerating scientific discovery.
When Large Multimodal Models Confront Evolving Knowledge:Challenges and Pathways
May 30, 2025Abstract:Large language/multimodal models (LLMs/LMMs) store extensive pre-trained knowledge but struggle to maintain consistency with real-world updates, making it difficult to avoid catastrophic forgetting while acquiring evolving knowledge. Previous work focused on constructing textual knowledge datasets and exploring knowledge injection in LLMs, lacking exploration of multimodal evolving knowledge injection in LMMs. To address this, we propose the EVOKE benchmark to evaluate LMMs' ability to inject multimodal evolving knowledge in real-world scenarios. Meanwhile, a comprehensive evaluation of multimodal evolving knowledge injection revealed two challenges: (1) Existing knowledge injection methods perform terribly on evolving knowledge. (2) Supervised fine-tuning causes catastrophic forgetting, particularly instruction following ability is severely compromised. Additionally, we provide pathways and find that: (1) Text knowledge augmentation during the training phase improves performance, while image augmentation cannot achieve it. (2) Continual learning methods, especially Replay and MoELoRA, effectively mitigate forgetting. Our findings indicate that current knowledge injection methods have many limitations on evolving knowledge, which motivates further research on more efficient and stable knowledge injection methods.
Use as Many Surrogates as You Want: Selective Ensemble Attack to Unleash Transferability without Sacrificing Resource Efficiency
May 19, 2025Abstract:In surrogate ensemble attacks, using more surrogate models yields higher transferability but lower resource efficiency. This practical trade-off between transferability and efficiency has largely limited existing attacks despite many pre-trained models are easily accessible online. In this paper, we argue that such a trade-off is caused by an unnecessary common assumption, i.e., all models should be identical across iterations. By lifting this assumption, we can use as many surrogates as we want to unleash transferability without sacrificing efficiency. Concretely, we propose Selective Ensemble Attack (SEA), which dynamically selects diverse models (from easily accessible pre-trained models) across iterations based on our new interpretation of decoupling within-iteration and cross-iteration model diversity.In this way, the number of within-iteration models is fixed for maintaining efficiency, while only cross-iteration model diversity is increased for higher transferability. Experiments on ImageNet demonstrate the superiority of SEA in various scenarios. For example, when dynamically selecting 4 from 20 accessible models, SEA yields 8.5% higher transferability than existing attacks under the same efficiency. The superiority of SEA also generalizes to real-world systems, such as commercial vision APIs and large vision-language models. Overall, SEA opens up the possibility of adaptively balancing transferability and efficiency according to specific resource requirements.
Improving Adversarial Transferability on Vision Transformers via Forward Propagation Refinement
Mar 19, 2025Abstract:Vision Transformers (ViTs) have been widely applied in various computer vision and vision-language tasks. To gain insights into their robustness in practical scenarios, transferable adversarial examples on ViTs have been extensively studied. A typical approach to improving adversarial transferability is by refining the surrogate model. However, existing work on ViTs has restricted their surrogate refinement to backward propagation. In this work, we instead focus on Forward Propagation Refinement (FPR) and specifically refine two key modules of ViTs: attention maps and token embeddings. For attention maps, we propose Attention Map Diversification (AMD), which diversifies certain attention maps and also implicitly imposes beneficial gradient vanishing during backward propagation. For token embeddings, we propose Momentum Token Embedding (MTE), which accumulates historical token embeddings to stabilize the forward updates in both the Attention and MLP blocks. We conduct extensive experiments with adversarial examples transferred from ViTs to various CNNs and ViTs, demonstrating that our FPR outperforms the current best (backward) surrogate refinement by up to 7.0\% on average. We also validate its superiority against popular defenses and its compatibility with other transfer methods. Codes and appendix are available at https://github.com/RYC-98/FPR.
Biology Instructions: A Dataset and Benchmark for Multi-Omics Sequence Understanding Capability of Large Language Models
Dec 26, 2024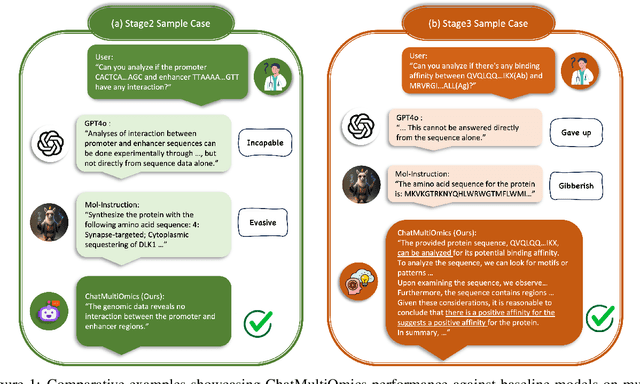
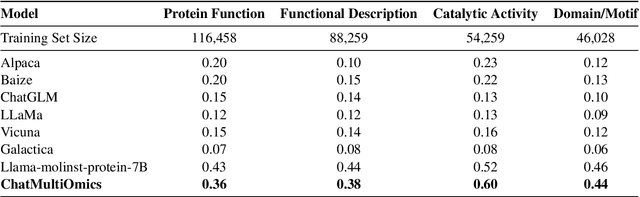
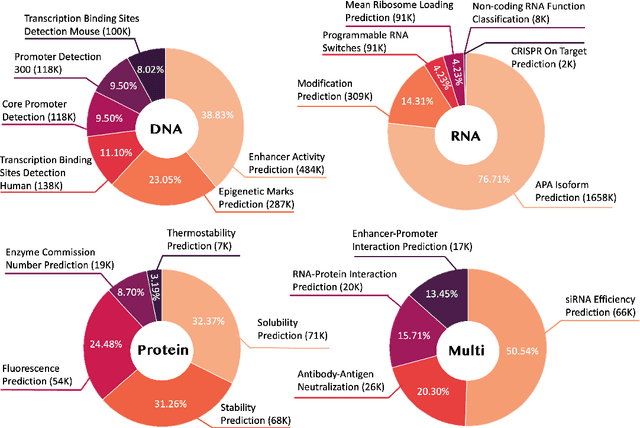
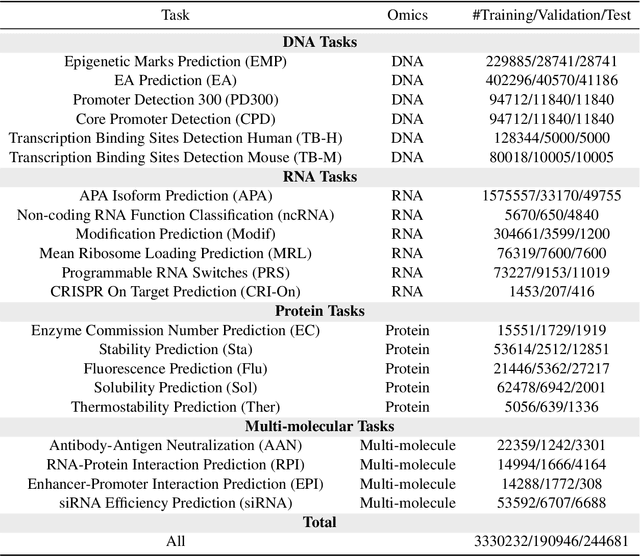
Abstract:Large language models have already demonstrated their formidable capabilities in general domains, ushering in a revolutionary transformation. However, exploring and exploiting the extensive knowledge of these models to comprehend multi-omics biology remains underexplored. To fill this research gap, we first introduce Biology-Instructions, the first large-scale multi-omics biological sequences-related instruction-tuning dataset including DNA, RNA, proteins, and multi-molecules, designed to bridge the gap between large language models (LLMs) and complex biological sequences-related tasks. This dataset can enhance the versatility of LLMs by integrating diverse biological sequenced-based prediction tasks with advanced reasoning capabilities, while maintaining conversational fluency. Additionally, we reveal significant performance limitations in even state-of-the-art LLMs on biological sequence-related multi-omics tasks without specialized pre-training and instruction-tuning. We further develop a strong baseline called ChatMultiOmics with a novel three-stage training pipeline, demonstrating the powerful ability to understand biology by using Biology-Instructions. Biology-Instructions and ChatMultiOmics are publicly available and crucial resources for enabling more effective integration of LLMs with multi-omics sequence analysis.
Improving Integrated Gradient-based Transferable Adversarial Examples by Refining the Integration Path
Dec 25, 2024



Abstract:Transferable adversarial examples are known to cause threats in practical, black-box attack scenarios. A notable approach to improving transferability is using integrated gradients (IG), originally developed for model interpretability. In this paper, we find that existing IG-based attacks have limited transferability due to their naive adoption of IG in model interpretability. To address this limitation, we focus on the IG integration path and refine it in three aspects: multiplicity, monotonicity, and diversity, supported by theoretical analyses. We propose the Multiple Monotonic Diversified Integrated Gradients (MuMoDIG) attack, which can generate highly transferable adversarial examples on different CNN and ViT models and defenses. Experiments validate that MuMoDIG outperforms the latest IG-based attack by up to 37.3\% and other state-of-the-art attacks by 8.4\%. In general, our study reveals that migrating established techniques to improve transferability may require non-trivial efforts. Code is available at \url{https://github.com/RYC-98/MuMoDIG}.
Model Decides How to Tokenize: Adaptive DNA Sequence Tokenization with MxDNA
Dec 18, 2024Abstract:Foundation models have made significant strides in understanding the genomic language of DNA sequences. However, previous models typically adopt the tokenization methods designed for natural language, which are unsuitable for DNA sequences due to their unique characteristics. In addition, the optimal approach to tokenize DNA remains largely under-explored, and may not be intuitively understood by humans even if discovered. To address these challenges, we introduce MxDNA, a novel framework where the model autonomously learns an effective DNA tokenization strategy through gradient decent. MxDNA employs a sparse Mixture of Convolution Experts coupled with a deformable convolution to model the tokenization process, with the discontinuous, overlapping, and ambiguous nature of meaningful genomic segments explicitly considered. On Nucleotide Transformer Benchmarks and Genomic Benchmarks, MxDNA demonstrates superior performance to existing methods with less pretraining data and time, highlighting its effectiveness. Finally, we show that MxDNA learns unique tokenization strategy distinct to those of previous methods and captures genomic functionalities at a token level during self-supervised pretraining. Our MxDNA aims to provide a new perspective on DNA tokenization, potentially offering broad applications in various domains and yielding profound insights.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge