Chunhua Weng
CPGPrompt: Translating Clinical Guidelines into LLM-Executable Decision Support
Jan 07, 2026Abstract:Clinical practice guidelines (CPGs) provide evidence-based recommendations for patient care; however, integrating them into Artificial Intelligence (AI) remains challenging. Previous approaches, such as rule-based systems, face significant limitations, including poor interpretability, inconsistent adherence to guidelines, and narrow domain applicability. To address this, we develop and validate CPGPrompt, an auto-prompting system that converts narrative clinical guidelines into large language models (LLMs). Our framework translates CPGs into structured decision trees and utilizes an LLM to dynamically navigate them for patient case evaluation. Synthetic vignettes were generated across three domains (headache, lower back pain, and prostate cancer) and distributed into four categories to test different decision scenarios. System performance was assessed on both binary specialty-referral decisions and fine-grained pathway-classification tasks. The binary specialty referral classification achieved consistently strong performance across all domains (F1: 0.85-1.00), with high recall (1.00 $\pm$ 0.00). In contrast, multi-class pathway assignment showed reduced performance, with domain-specific variations: headache (F1: 0.47), lower back pain (F1: 0.72), and prostate cancer (F1: 0.77). Domain-specific performance differences reflected the structure of each guideline. The headache guideline highlighted challenges with negation handling. The lower back pain guideline required temporal reasoning. In contrast, prostate cancer pathways benefited from quantifiable laboratory tests, resulting in more reliable decision-making.
Toward Global Large Language Models in Medicine
Jan 05, 2026Abstract:Despite continuous advances in medical technology, the global distribution of health care resources remains uneven. The development of large language models (LLMs) has transformed the landscape of medicine and holds promise for improving health care quality and expanding access to medical information globally. However, existing LLMs are primarily trained on high-resource languages, limiting their applicability in global medical scenarios. To address this gap, we constructed GlobMed, a large multilingual medical dataset, containing over 500,000 entries spanning 12 languages, including four low-resource languages. Building on this, we established GlobMed-Bench, which systematically assesses 56 state-of-the-art proprietary and open-weight LLMs across multiple multilingual medical tasks, revealing significant performance disparities across languages, particularly for low-resource languages. Additionally, we introduced GlobMed-LLMs, a suite of multilingual medical LLMs trained on GlobMed, with parameters ranging from 1.7B to 8B. GlobMed-LLMs achieved an average performance improvement of over 40% relative to baseline models, with a more than threefold increase in performance on low-resource languages. Together, these resources provide an important foundation for advancing the equitable development and application of LLMs globally, enabling broader language communities to benefit from technological advances.
A Method for Characterizing Disease Progression from Acute Kidney Injury to Chronic Kidney Disease
Nov 18, 2025Abstract:Patients with acute kidney injury (AKI) are at high risk of developing chronic kidney disease (CKD), but identifying those at greatest risk remains challenging. We used electronic health record (EHR) data to dynamically track AKI patients' clinical evolution and characterize AKI-to-CKD progression. Post-AKI clinical states were identified by clustering patient vectors derived from longitudinal medical codes and creatinine measurements. Transition probabilities between states and progression to CKD were estimated using multi-state modeling. After identifying common post-AKI trajectories, CKD risk factors in AKI subpopulations were identified through survival analysis. Of 20,699 patients with AKI at admission, 3,491 (17%) developed CKD. We identified fifteen distinct post-AKI states, each with different probabilities of CKD development. Most patients (75%, n=15,607) remained in a single state or made only one transition during the study period. Both established (e.g., AKI severity, diabetes, hypertension, heart failure, liver disease) and novel CKD risk factors, with their impact varying across these clinical states. This study demonstrates a data-driven approach for identifying high-risk AKI patients, supporting the development of decision-support tools for early CKD detection and intervention.
Natural Language Processing in Support of Evidence-based Medicine: A Scoping Review
May 28, 2025


Abstract:Evidence-based medicine (EBM) is at the forefront of modern healthcare, emphasizing the use of the best available scientific evidence to guide clinical decisions. Due to the sheer volume and rapid growth of medical literature and the high cost of curation, there is a critical need to investigate Natural Language Processing (NLP) methods to identify, appraise, synthesize, summarize, and disseminate evidence in EBM. This survey presents an in-depth review of 129 research studies on leveraging NLP for EBM, illustrating its pivotal role in enhancing clinical decision-making processes. The paper systematically explores how NLP supports the five fundamental steps of EBM -- Ask, Acquire, Appraise, Apply, and Assess. The review not only identifies current limitations within the field but also proposes directions for future research, emphasizing the potential for NLP to revolutionize EBM by refining evidence extraction, evidence synthesis, appraisal, summarization, enhancing data comprehensibility, and facilitating a more efficient clinical workflow.
Machine Learning Applications Related to Suicide in Military and Veterans: A Scoping Literature Review
May 18, 2025Abstract:Suicide remains one of the main preventable causes of death among active service members and veterans. Early detection and prediction are crucial in suicide prevention. Machine learning techniques have yielded promising results in this area recently. This study aims to assess and summarize current research and provides a comprehensive review regarding the application of machine learning techniques in assessing and predicting suicidal ideation, attempts, and mortality among members of military and veteran populations. A keyword search using PubMed, IEEE, ACM, and Google Scholar was conducted, and the PRISMA protocol was adopted for relevant study selection. Thirty-two articles met the inclusion criteria. These studies consistently identified risk factors relevant to mental health issues such as depression, post-traumatic stress disorder (PTSD), suicidal ideation, prior attempts, physical health problems, and demographic characteristics. Machine learning models applied in this area have demonstrated reasonable predictive accuracy. However, additional research gaps still exist. First, many studies have overlooked metrics that distinguish between false positives and negatives, such as positive predictive value and negative predictive value, which are crucial in the context of suicide prevention policies. Second, more dedicated approaches to handling survival and longitudinal data should be explored. Lastly, most studies focused on machine learning methods, with limited discussion of their connection to clinical rationales. In summary, machine learning analyses have identified a wide range of risk factors associated with suicide in military populations. The diversity and complexity of these factors also demonstrates that effective prevention strategies must be comprehensive and flexible.
WatchGuardian: Enabling User-Defined Personalized Just-in-Time Intervention on Smartwatch
Feb 09, 2025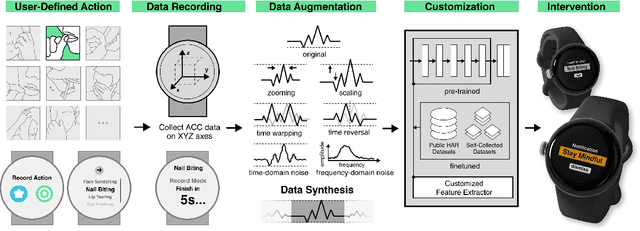
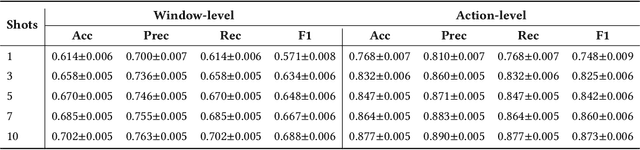
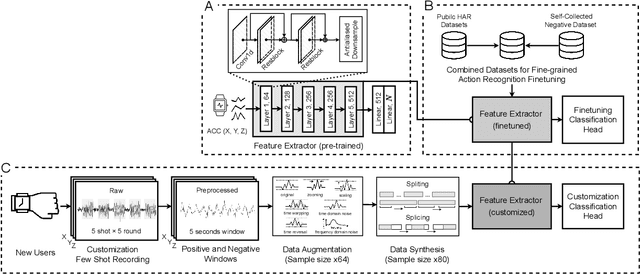
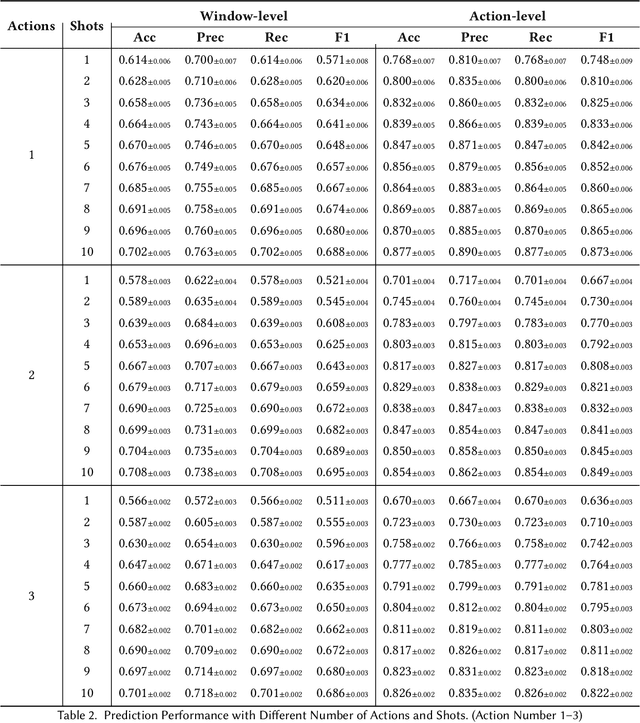
Abstract:While just-in-time interventions (JITIs) have effectively targeted common health behaviors, individuals often have unique needs to intervene in personal undesirable actions that can negatively affect physical, mental, and social well-being. We present WatchGuardian, a smartwatch-based JITI system that empowers users to define custom interventions for these personal actions with a small number of samples. For the model to detect new actions based on limited new data samples, we developed a few-shot learning pipeline that finetuned a pre-trained inertial measurement unit (IMU) model on public hand-gesture datasets. We then designed a data augmentation and synthesis process to train additional classification layers for customization. Our offline evaluation with 26 participants showed that with three, five, and ten examples, our approach achieved an average accuracy of 76.8%, 84.7%, and 87.7%, and an F1 score of 74.8%, 84.2%, and 87.2% We then conducted a four-hour intervention study to compare WatchGuardian against a rule-based intervention. Our results demonstrated that our system led to a significant reduction by 64.0 +- 22.6% in undesirable actions, substantially outperforming the baseline by 29.0%. Our findings underscore the effectiveness of a customizable, AI-driven JITI system for individuals in need of behavioral intervention in personal undesirable actions. We envision that our work can inspire broader applications of user-defined personalized intervention with advanced AI solutions.
Semi-Supervised Learning from Small Annotated Data and Large Unlabeled Data for Fine-grained PICO Entity Recognition
Dec 26, 2024



Abstract:Objective: Extracting PICO elements -- Participants, Intervention, Comparison, and Outcomes -- from clinical trial literature is essential for clinical evidence retrieval, appraisal, and synthesis. Existing approaches do not distinguish the attributes of PICO entities. This study aims to develop a named entity recognition (NER) model to extract PICO entities with fine granularities. Materials and Methods: Using a corpus of 2,511 abstracts with PICO mentions from 4 public datasets, we developed a semi-supervised method to facilitate the training of a NER model, FinePICO, by combining limited annotated data of PICO entities and abundant unlabeled data. For evaluation, we divided the entire dataset into two subsets: a smaller group with annotations and a larger group without annotations. We then established the theoretical lower and upper performance bounds based on the performance of supervised learning models trained solely on the small, annotated subset and on the entire set with complete annotations, respectively. Finally, we evaluated FinePICO on both the smaller annotated subset and the larger, initially unannotated subset. We measured the performance of FinePICO using precision, recall, and F1. Results: Our method achieved precision/recall/F1 of 0.567/0.636/0.60, respectively, using a small set of annotated samples, outperforming the baseline model (F1: 0.437) by more than 16\%. The model demonstrates generalizability to a different PICO framework and to another corpus, which consistently outperforms the benchmark in diverse experimental settings (p-value \textless0.001). Conclusion: This study contributes a generalizable and effective semi-supervised approach to named entity recognition leveraging large unlabeled data together with small, annotated data. It also initially supports fine-grained PICO extraction.
A MapReduce Approach to Effectively Utilize Long Context Information in Retrieval Augmented Language Models
Dec 17, 2024



Abstract:While holding great promise for improving and facilitating healthcare, large language models (LLMs) struggle to produce up-to-date responses on evolving topics due to outdated knowledge or hallucination. Retrieval-augmented generation (RAG) is a pivotal innovation that improves the accuracy and relevance of LLM responses by integrating LLMs with a search engine and external sources of knowledge. However, the quality of RAG responses can be largely impacted by the rank and density of key information in the retrieval results, such as the "lost-in-the-middle" problem. In this work, we aim to improve the robustness and reliability of the RAG workflow in the medical domain. Specifically, we propose a map-reduce strategy, BriefContext, to combat the "lost-in-the-middle" issue without modifying the model weights. We demonstrated the advantage of the workflow with various LLM backbones and on multiple QA datasets. This method promises to improve the safety and reliability of LLMs deployed in healthcare domains.
Demystifying Large Language Models for Medicine: A Primer
Oct 24, 2024



Abstract:Large language models (LLMs) represent a transformative class of AI tools capable of revolutionizing various aspects of healthcare by generating human-like responses across diverse contexts and adapting to novel tasks following human instructions. Their potential application spans a broad range of medical tasks, such as clinical documentation, matching patients to clinical trials, and answering medical questions. In this primer paper, we propose an actionable guideline to help healthcare professionals more efficiently utilize LLMs in their work, along with a set of best practices. This approach consists of several main phases, including formulating the task, choosing LLMs, prompt engineering, fine-tuning, and deployment. We start with the discussion of critical considerations in identifying healthcare tasks that align with the core capabilities of LLMs and selecting models based on the selected task and data, performance requirements, and model interface. We then review the strategies, such as prompt engineering and fine-tuning, to adapt standard LLMs to specialized medical tasks. Deployment considerations, including regulatory compliance, ethical guidelines, and continuous monitoring for fairness and bias, are also discussed. By providing a structured step-by-step methodology, this tutorial aims to equip healthcare professionals with the tools necessary to effectively integrate LLMs into clinical practice, ensuring that these powerful technologies are applied in a safe, reliable, and impactful manner.
Closing the gap between open-source and commercial large language models for medical evidence summarization
Jul 25, 2024



Abstract:Large language models (LLMs) hold great promise in summarizing medical evidence. Most recent studies focus on the application of proprietary LLMs. Using proprietary LLMs introduces multiple risk factors, including a lack of transparency and vendor dependency. While open-source LLMs allow better transparency and customization, their performance falls short compared to proprietary ones. In this study, we investigated to what extent fine-tuning open-source LLMs can further improve their performance in summarizing medical evidence. Utilizing a benchmark dataset, MedReview, consisting of 8,161 pairs of systematic reviews and summaries, we fine-tuned three broadly-used, open-sourced LLMs, namely PRIMERA, LongT5, and Llama-2. Overall, the fine-tuned LLMs obtained an increase of 9.89 in ROUGE-L (95% confidence interval: 8.94-10.81), 13.21 in METEOR score (95% confidence interval: 12.05-14.37), and 15.82 in CHRF score (95% confidence interval: 13.89-16.44). The performance of fine-tuned LongT5 is close to GPT-3.5 with zero-shot settings. Furthermore, smaller fine-tuned models sometimes even demonstrated superior performance compared to larger zero-shot models. The above trends of improvement were also manifested in both human and GPT4-simulated evaluations. Our results can be applied to guide model selection for tasks demanding particular domain knowledge, such as medical evidence summarization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge