Guotong Xie
IAPT: Instruction-Aware Prompt Tuning for Large Language Models
May 28, 2024



Abstract:Soft prompt tuning is a widely studied parameter-efficient fine-tuning method. However, it has a clear drawback: many soft tokens must be inserted into the input sequences to guarantee downstream performance. As a result, soft prompt tuning is less considered than Low-rank adaptation (LoRA) in the large language modeling (LLM) era. In this work, we propose a novel prompt tuning method, Instruction-Aware Prompt Tuning (IAPT), that requires only four soft tokens. First, we install a parameter-efficient soft prompt generator at each Transformer layer to generate idiosyncratic soft prompts for each input instruction. The generated soft prompts can be seen as a semantic summary of the input instructions and can effectively guide the output generation. Second, the soft prompt generators are modules with a bottleneck architecture consisting of a self-attention pooling operation, two linear projections, and an activation function. Pilot experiments show that prompt generators at different Transformer layers require different activation functions. Thus, we propose to learn the idiosyncratic activation functions for prompt generators automatically with the help of rational functions. We have conducted experiments on various tasks, and the experimental results demonstrate that (a) our IAPT method can outperform the recent baselines with comparable tunable parameters. (b) Our IAPT method is more efficient than LoRA under the single-backbone multi-tenant setting.
Text2MDT: Extracting Medical Decision Trees from Medical Texts
Jan 04, 2024Abstract:Knowledge of the medical decision process, which can be modeled as medical decision trees (MDTs), is critical to build clinical decision support systems. However, the current MDT construction methods rely heavily on time-consuming and laborious manual annotation. In this work, we propose a novel task, Text2MDT, to explore the automatic extraction of MDTs from medical texts such as medical guidelines and textbooks. We normalize the form of the MDT and create an annotated Text-to-MDT dataset in Chinese with the participation of medical experts. We investigate two different methods for the Text2MDT tasks: (a) an end-to-end framework which only relies on a GPT style large language models (LLM) instruction tuning to generate all the node information and tree structures. (b) The pipeline framework which decomposes the Text2MDT task to three subtasks. Experiments on our Text2MDT dataset demonstrate that: (a) the end-to-end method basd on LLMs (7B parameters or larger) show promising results, and successfully outperform the pipeline methods. (b) The chain-of-thought (COT) prompting method \cite{Wei2022ChainOT} can improve the performance of the fine-tuned LLMs on the Text2MDT test set. (c) the lightweight pipelined method based on encoder-based pretrained models can perform comparably with LLMs with model complexity two magnititudes smaller. Our Text2MDT dataset is open-sourced at \url{https://tianchi.aliyun.com/dataset/95414}, and the source codes are open-sourced at \url{https://github.com/michael-wzhu/text2dt}.
UltraFeedback: Boosting Language Models with High-quality Feedback
Oct 02, 2023



Abstract:Reinforcement learning from human feedback (RLHF) has become a pivot technique in aligning large language models (LLMs) with human preferences. In RLHF practice, preference data plays a crucial role in bridging human proclivity and LLMs. However, the scarcity of diverse, naturalistic datasets of human preferences on LLM outputs at scale poses a great challenge to RLHF as well as feedback learning research within the open-source community. Current preference datasets, either proprietary or limited in size and prompt variety, result in limited RLHF adoption in open-source models and hinder further exploration. In this study, we propose ULTRAFEEDBACK, a large-scale, high-quality, and diversified preference dataset designed to overcome these limitations and foster RLHF development. To create ULTRAFEEDBACK, we compile a diverse array of instructions and models from multiple sources to produce comparative data. We meticulously devise annotation instructions and employ GPT-4 to offer detailed feedback in both numerical and textual forms. ULTRAFEEDBACK establishes a reproducible and expandable preference data construction pipeline, serving as a solid foundation for future RLHF and feedback learning research. Utilizing ULTRAFEEDBACK, we train various models to demonstrate its effectiveness, including the reward model UltraRM, chat language model UltraLM-13B-PPO, and critique model UltraCM. Experimental results indicate that our models outperform existing open-source models, achieving top performance across multiple benchmarks. Our data and models are available at https://github.com/thunlp/UltraFeedback.
Interactive Molecular Discovery with Natural Language
Jun 21, 2023Abstract:Natural language is expected to be a key medium for various human-machine interactions in the era of large language models. When it comes to the biochemistry field, a series of tasks around molecules (e.g., property prediction, molecule mining, etc.) are of great significance while having a high technical threshold. Bridging the molecule expressions in natural language and chemical language can not only hugely improve the interpretability and reduce the operation difficulty of these tasks, but also fuse the chemical knowledge scattered in complementary materials for a deeper comprehension of molecules. Based on these benefits, we propose the conversational molecular design, a novel task adopting natural language for describing and editing target molecules. To better accomplish this task, we design ChatMol, a knowledgeable and versatile generative pre-trained model, enhanced by injecting experimental property information, molecular spatial knowledge, and the associations between natural and chemical languages into it. Several typical solutions including large language models (e.g., ChatGPT) are evaluated, proving the challenge of conversational molecular design and the effectiveness of our knowledge enhancement method. Case observations and analysis are conducted to provide directions for further exploration of natural-language interaction in molecular discovery.
Unified Demonstration Retriever for In-Context Learning
May 16, 2023



Abstract:In-context learning is a new learning paradigm where a language model conditions on a few input-output pairs (demonstrations) and a test input, and directly outputs the prediction. It has been shown highly dependent on the provided demonstrations and thus promotes the research of demonstration retrieval: given a test input, relevant examples are retrieved from the training set to serve as informative demonstrations for in-context learning. While previous works focus on training task-specific retrievers for several tasks separately, these methods are often hard to transfer and scale on various tasks, and separately trained retrievers incur a lot of parameter storage and deployment cost. In this paper, we propose Unified Demonstration Retriever (\textbf{UDR}), a single model to retrieve demonstrations for a wide range of tasks. To train UDR, we cast various tasks' training signals into a unified list-wise ranking formulation by language model's feedback. Then we propose a multi-task list-wise ranking training framework, with an iterative mining strategy to find high-quality candidates, which can help UDR fully incorporate various tasks' signals. Experiments on 30+ tasks across 13 task families and multiple data domains show that UDR significantly outperforms baselines. Further analyses show the effectiveness of each proposed component and UDR's strong ability in various scenarios including different LMs (1.3B - 175B), unseen datasets, varying demonstration quantities, etc.
Filter Pruning via Filters Similarity in Consecutive Layers
Apr 26, 2023Abstract:Filter pruning is widely adopted to compress and accelerate the Convolutional Neural Networks (CNNs), but most previous works ignore the relationship between filters and channels in different layers. Processing each layer independently fails to utilize the collaborative relationship across layers. In this paper, we intuitively propose a novel pruning method by explicitly leveraging the Filters Similarity in Consecutive Layers (FSCL). FSCL compresses models by pruning filters whose corresponding features are more worthless in the model. The extensive experiments demonstrate the effectiveness of FSCL, and it yields remarkable improvement over state-of-the-art on accuracy, FLOPs and parameter reduction on several benchmark models and datasets.
SFE-AI at SemEval-2022 Task 11: Low-Resource Named Entity Recognition using Large Pre-trained Language Models
May 29, 2022
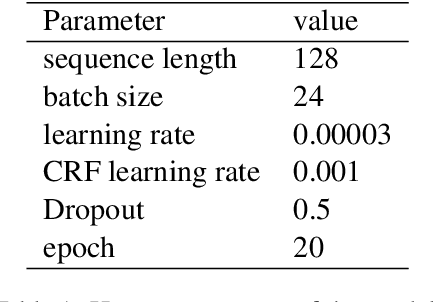
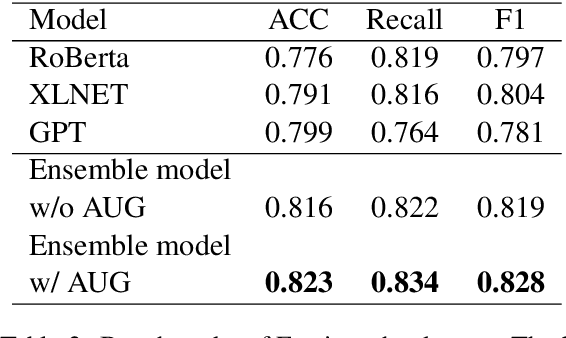
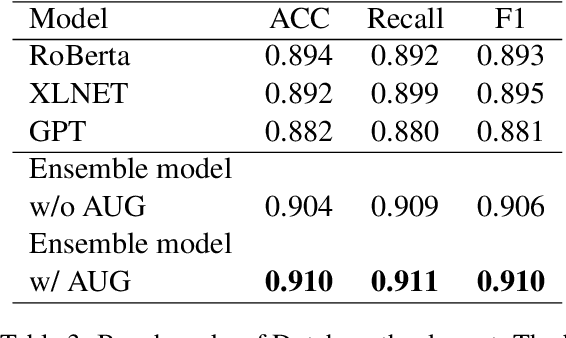
Abstract:Large scale pre-training models have been widely used in named entity recognition (NER) tasks. However, model ensemble through parameter averaging or voting can not give full play to the differentiation advantages of different models, especially in the open domain. This paper describes our NER system in the SemEval 2022 task11: MultiCoNER. We proposed an effective system to adaptively ensemble pre-trained language models by a Transformer layer. By assigning different weights to each model for different inputs, we adopted the Transformer layer to integrate the advantages of diverse models effectively. Experimental results show that our method achieves superior performances in Farsi and Dutch.
PASH at TREC 2021 Deep Learning Track: Generative Enhanced Model for Multi-stage Ranking
May 24, 2022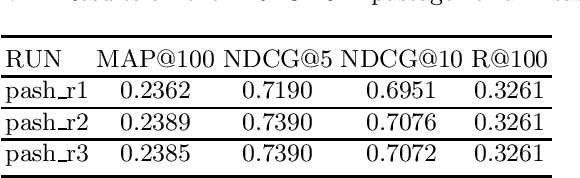
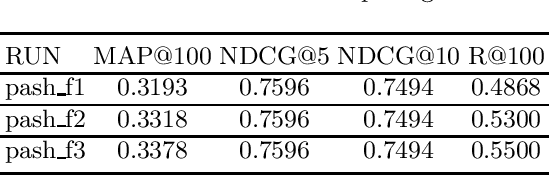
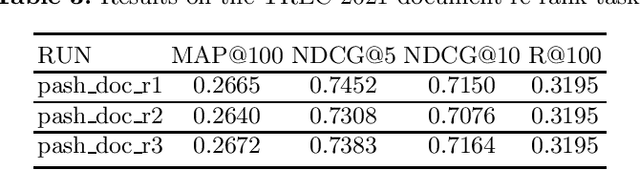
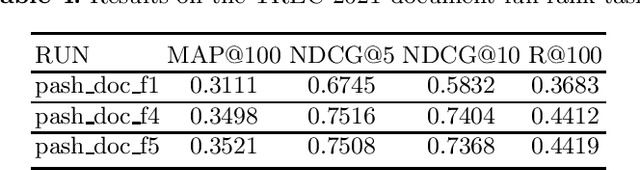
Abstract:This paper describes the PASH participation in TREC 2021 Deep Learning Track. In the recall stage, we adopt a scheme combining sparse and dense retrieval method. In the multi-stage ranking phase, point-wise and pair-wise ranking strategies are used one after another based on model continual pre-trained on general knowledge and document-level data. Compared to TREC 2020 Deep Learning Track, we have additionally introduced the generative model T5 to further enhance the performance.
Automatic Fine-grained Glomerular Lesion Recognition in Kidney Pathology
Mar 11, 2022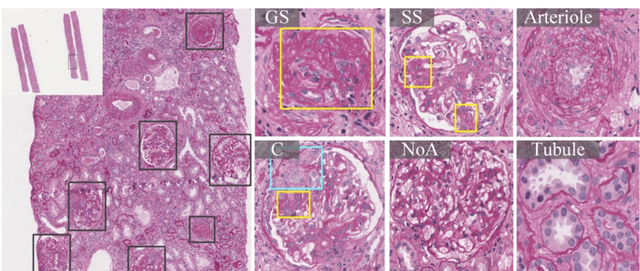
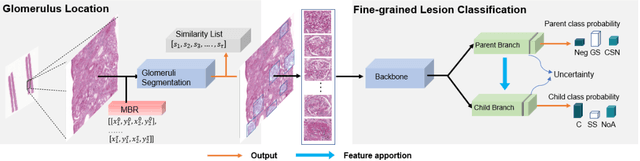
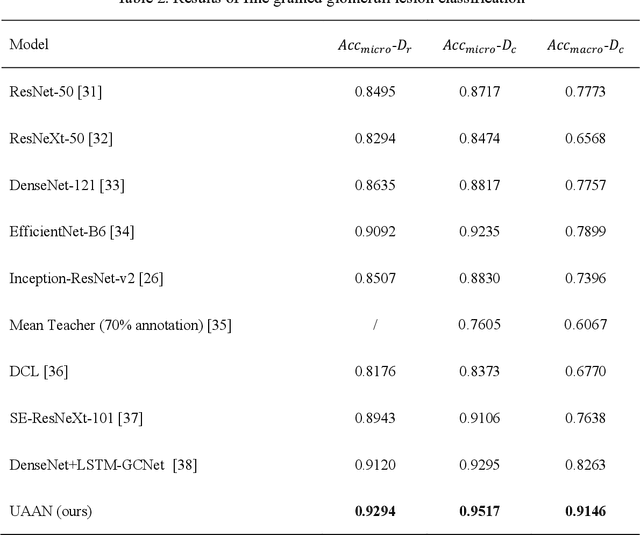
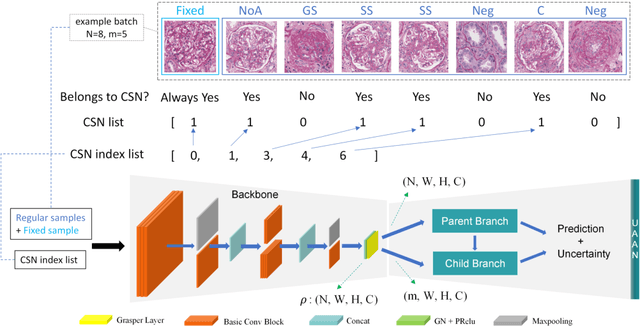
Abstract:Recognition of glomeruli lesions is the key for diagnosis and treatment planning in kidney pathology; however, the coexisting glomerular structures such as mesangial regions exacerbate the difficulties of this task. In this paper, we introduce a scheme to recognize fine-grained glomeruli lesions from whole slide images. First, a focal instance structural similarity loss is proposed to drive the model to locate all types of glomeruli precisely. Then an Uncertainty Aided Apportionment Network is designed to carry out the fine-grained visual classification without bounding-box annotations. This double branch-shaped structure extracts common features of the child class from the parent class and produces the uncertainty factor for reconstituting the training dataset. Results of slide-wise evaluation illustrate the effectiveness of the entire scheme, with an 8-22% improvement of the mean Average Precision compared with remarkable detection methods. The comprehensive results clearly demonstrate the effectiveness of the proposed method.
A Simple Hash-Based Early Exiting Approach For Language Understanding and Generation
Mar 03, 2022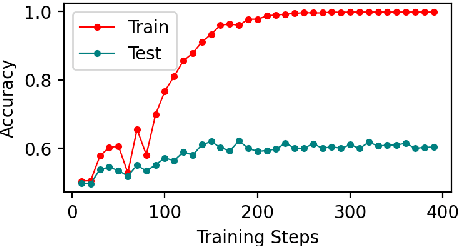
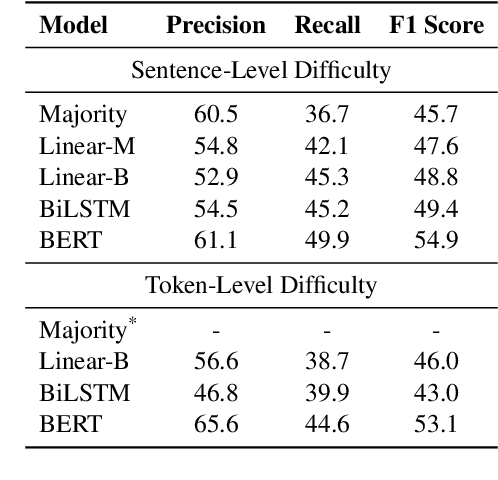
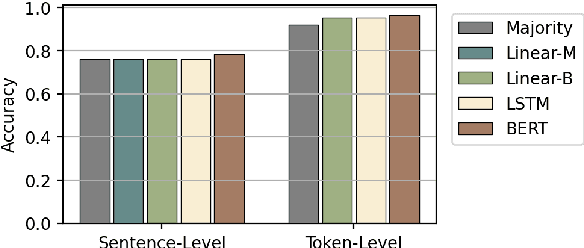
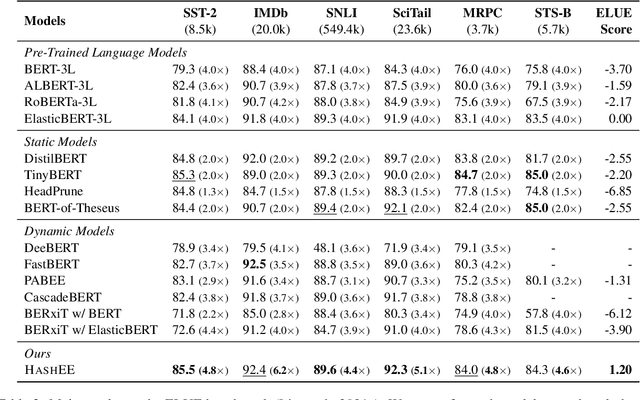
Abstract:Early exiting allows instances to exit at different layers according to the estimation of difficulty. Previous works usually adopt heuristic metrics such as the entropy of internal outputs to measure instance difficulty, which suffers from generalization and threshold-tuning. In contrast, learning to exit, or learning to predict instance difficulty is a more appealing way. Though some effort has been devoted to employing such "learn-to-exit" modules, it is still unknown whether and how well the instance difficulty can be learned. As a response, we first conduct experiments on the learnability of instance difficulty, which demonstrates that modern neural models perform poorly on predicting instance difficulty. Based on this observation, we propose a simple-yet-effective Hash-based Early Exiting approach (HashEE) that replaces the learn-to-exit modules with hash functions to assign each token to a fixed exiting layer. Different from previous methods, HashEE requires no internal classifiers nor extra parameters, and therefore is more efficient. Experimental results on classification, regression, and generation tasks demonstrate that HashEE can achieve higher performance with fewer FLOPs and inference time compared with previous state-of-the-art early exiting methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge