Chang Yu
MergeDNA: Context-aware Genome Modeling with Dynamic Tokenization through Token Merging
Nov 17, 2025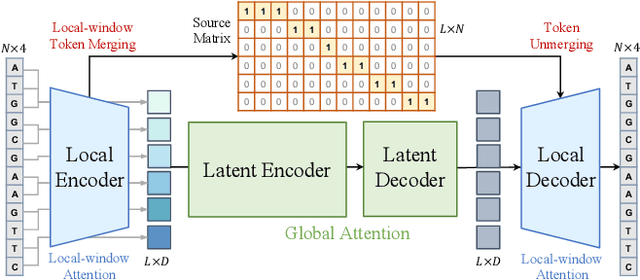
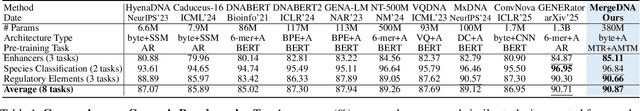
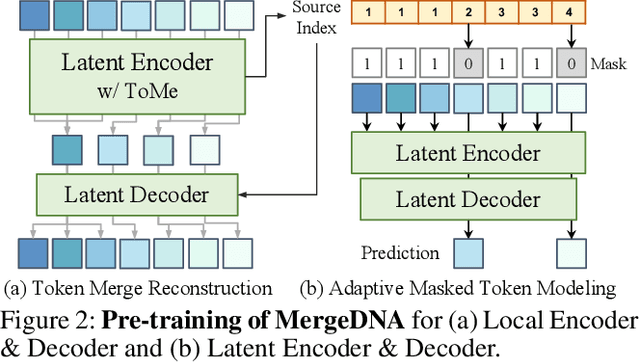
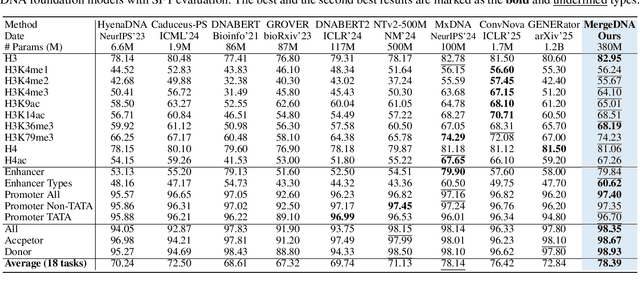
Abstract:Modeling genomic sequences faces two unsolved challenges: the information density varies widely across different regions, while there is no clearly defined minimum vocabulary unit. Relying on either four primitive bases or independently designed DNA tokenizers, existing approaches with naive masked language modeling pre-training often fail to adapt to the varying complexities of genomic sequences. Leveraging Token Merging techniques, this paper introduces a hierarchical architecture that jointly optimizes a dynamic genomic tokenizer and latent Transformers with context-aware pre-training tasks. As for network structures, the tokenization module automatically chunks adjacent bases into words by stacking multiple layers of the differentiable token merging blocks with local-window constraints, then a Latent Encoder captures the global context of these merged words by full-attention blocks. Symmetrically employing a Latent Decoder and a Local Decoder, MergeDNA learns with two pre-training tasks: Merged Token Reconstruction simultaneously trains the dynamic tokenization module and adaptively filters important tokens, while Adaptive Masked Token Modeling learns to predict these filtered tokens to capture informative contents. Extensive experiments show that MergeDNA achieves superior performance on three popular DNA benchmarks and several multi-omics tasks with fine-tuning or zero-shot evaluation, outperforming typical tokenization methods and large-scale DNA foundation models.
Right-Side-Out: Learning Zero-Shot Sim-to-Real Garment Reversal
Sep 19, 2025Abstract:Turning garments right-side out is a challenging manipulation task: it is highly dynamic, entails rapid contact changes, and is subject to severe visual occlusion. We introduce Right-Side-Out, a zero-shot sim-to-real framework that effectively solves this challenge by exploiting task structures. We decompose the task into Drag/Fling to create and stabilize an access opening, followed by Insert&Pull to invert the garment. Each step uses a depth-inferred, keypoint-parameterized bimanual primitive that sharply reduces the action space while preserving robustness. Efficient data generation is enabled by our custom-built, high-fidelity, GPU-parallel Material Point Method (MPM) simulator that models thin-shell deformation and provides robust and efficient contact handling for batched rollouts. Built on the simulator, our fully automated pipeline scales data generation by randomizing garment geometry, material parameters, and viewpoints, producing depth, masks, and per-primitive keypoint labels without any human annotations. With a single depth camera, policies trained entirely in simulation deploy zero-shot on real hardware, achieving up to 81.3% success rate. By employing task decomposition and high fidelity simulation, our framework enables tackling highly dynamic, severely occluded tasks without laborious human demonstrations.
EvoCoT: Overcoming the Exploration Bottleneck in Reinforcement Learning
Aug 11, 2025Abstract:Reinforcement learning with verifiable reward (RLVR) has become a promising paradigm for post-training large language models (LLMs) to improve their reasoning capability. However, when the rollout accuracy is low on hard problems, the reward becomes sparse, limiting learning efficiency and causing exploration bottlenecks. Existing approaches either rely on stronger LLMs for distillation or filter out difficult problems, which limits scalability or restricts reasoning improvement through exploration. We propose EvoCoT, a self-evolving curriculum learning framework based on two-stage chain-of-thought (CoT) reasoning optimization. EvoCoT constrains the exploration space by self-generating and verifying CoT trajectories, then gradually shortens them to expand the space in a controlled way. This enables LLMs to stably learn from initially unsolved hard problems under sparse rewards. We apply EvoCoT to multiple LLM families, including Qwen, DeepSeek, and Llama. Experiments show that EvoCoT enables LLMs to solve previously unsolved problems, improves reasoning capability without external CoT supervision, and is compatible with various RL fine-tuning methods. We release the source code to support future research.
Revisiting Randomization in Greedy Model Search
Jun 18, 2025



Abstract:Combining randomized estimators in an ensemble, such as via random forests, has become a fundamental technique in modern data science, but can be computationally expensive. Furthermore, the mechanism by which this improves predictive performance is poorly understood. We address these issues in the context of sparse linear regression by proposing and analyzing an ensemble of greedy forward selection estimators that are randomized by feature subsampling -- at each iteration, the best feature is selected from within a random subset. We design a novel implementation based on dynamic programming that greatly improves its computational efficiency. Furthermore, we show via careful numerical experiments that our method can outperform popular methods such as lasso and elastic net across a wide range of settings. Next, contrary to prevailing belief that randomized ensembling is analogous to shrinkage, we show via numerical experiments that it can simultaneously reduce training error and degrees of freedom, thereby shifting the entire bias-variance trade-off curve of the base estimator. We prove this fact rigorously in the setting of orthogonal features, in which case, the ensemble estimator rescales the ordinary least squares coefficients with a two-parameter family of logistic weights, thereby enlarging the model search space. These results enhance our understanding of random forests and suggest that implicit regularization in general may have more complicated effects than explicit regularization.
Gradient Boosting Decision Tree with LSTM for Investment Prediction
May 29, 2025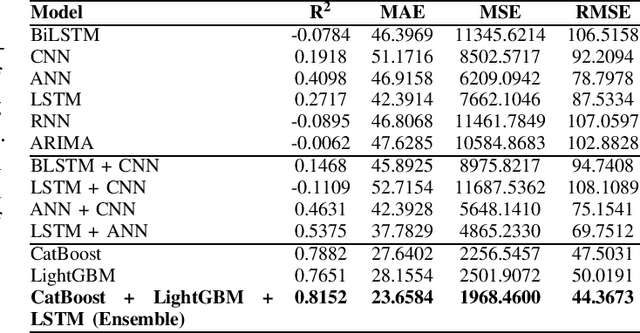
Abstract:This paper proposes a hybrid framework combining LSTM (Long Short-Term Memory) networks with LightGBM and CatBoost for stock price prediction. The framework processes time-series financial data and evaluates performance using seven models: Artificial Neural Networks (ANNs), Convolutional Neural Networks (CNNs), Bidirectional LSTM (BiLSTM), vanilla LSTM, XGBoost, LightGBM, and standard Neural Networks (NNs). Key metrics, including MAE, R-squared, MSE, and RMSE, are used to establish benchmarks across different time scales. Building on these benchmarks, we develop an ensemble model that combines the strengths of sequential and tree-based approaches. Experimental results show that the proposed framework improves accuracy by 10 to 15 percent compared to individual models and reduces error during market changes. This study highlights the potential of ensemble methods for financial forecasting and provides a flexible design for integrating new machine learning techniques.
GRAPE: Heterogeneous Graph Representation Learning for Genetic Perturbation with Coding and Non-Coding Biotype
May 06, 2025Abstract:Predicting genetic perturbations enables the identification of potentially crucial genes prior to wet-lab experiments, significantly improving overall experimental efficiency. Since genes are the foundation of cellular life, building gene regulatory networks (GRN) is essential to understand and predict the effects of genetic perturbations. However, current methods fail to fully leverage gene-related information, and solely rely on simple evaluation metrics to construct coarse-grained GRN. More importantly, they ignore functional differences between biotypes, limiting the ability to capture potential gene interactions. In this work, we leverage pre-trained large language model and DNA sequence model to extract features from gene descriptions and DNA sequence data, respectively, which serve as the initialization for gene representations. Additionally, we introduce gene biotype information for the first time in genetic perturbation, simulating the distinct roles of genes with different biotypes in regulating cellular processes, while capturing implicit gene relationships through graph structure learning (GSL). We propose GRAPE, a heterogeneous graph neural network (HGNN) that leverages gene representations initialized with features from descriptions and sequences, models the distinct roles of genes with different biotypes, and dynamically refines the GRN through GSL. The results on publicly available datasets show that our method achieves state-of-the-art performance.
Taccel: Scaling Up Vision-based Tactile Robotics via High-performance GPU Simulation
Apr 17, 2025Abstract:Tactile sensing is crucial for achieving human-level robotic capabilities in manipulation tasks. VBTSs have emerged as a promising solution, offering high spatial resolution and cost-effectiveness by sensing contact through camera-captured deformation patterns of elastic gel pads. However, these sensors' complex physical characteristics and visual signal processing requirements present unique challenges for robotic applications. The lack of efficient and accurate simulation tools for VBTS has significantly limited the scale and scope of tactile robotics research. Here we present Taccel, a high-performance simulation platform that integrates IPC and ABD to model robots, tactile sensors, and objects with both accuracy and unprecedented speed, achieving an 18-fold acceleration over real-time across thousands of parallel environments. Unlike previous simulators that operate at sub-real-time speeds with limited parallelization, Taccel provides precise physics simulation and realistic tactile signals while supporting flexible robot-sensor configurations through user-friendly APIs. Through extensive validation in object recognition, robotic grasping, and articulated object manipulation, we demonstrate precise simulation and successful sim-to-real transfer. These capabilities position Taccel as a powerful tool for scaling up tactile robotics research and development. By enabling large-scale simulation and experimentation with tactile sensing, Taccel accelerates the development of more capable robotic systems, potentially transforming how robots interact with and understand their physical environment.
Cross-Frequency Implicit Neural Representation with Self-Evolving Parameters
Apr 15, 2025Abstract:Implicit neural representation (INR) has emerged as a powerful paradigm for visual data representation. However, classical INR methods represent data in the original space mixed with different frequency components, and several feature encoding parameters (e.g., the frequency parameter $\omega$ or the rank $R$) need manual configurations. In this work, we propose a self-evolving cross-frequency INR using the Haar wavelet transform (termed CF-INR), which decouples data into four frequency components and employs INRs in the wavelet space. CF-INR allows the characterization of different frequency components separately, thus enabling higher accuracy for data representation. To more precisely characterize cross-frequency components, we propose a cross-frequency tensor decomposition paradigm for CF-INR with self-evolving parameters, which automatically updates the rank parameter $R$ and the frequency parameter $\omega$ for each frequency component through self-evolving optimization. This self-evolution paradigm eliminates the laborious manual tuning of these parameters, and learns a customized cross-frequency feature encoding configuration for each dataset. We evaluate CF-INR on a variety of visual data representation and recovery tasks, including image regression, inpainting, denoising, and cloud removal. Extensive experiments demonstrate that CF-INR outperforms state-of-the-art methods in each case.
MergeVQ: A Unified Framework for Visual Generation and Representation with Disentangled Token Merging and Quantization
Apr 01, 2025Abstract:Masked Image Modeling (MIM) with Vector Quantization (VQ) has achieved great success in both self-supervised pre-training and image generation. However, most existing methods struggle to address the trade-off in shared latent space for generation quality vs. representation learning and efficiency. To push the limits of this paradigm, we propose MergeVQ, which incorporates token merging techniques into VQ-based generative models to bridge the gap between image generation and visual representation learning in a unified architecture. During pre-training, MergeVQ decouples top-k semantics from latent space with the token merge module after self-attention blocks in the encoder for subsequent Look-up Free Quantization (LFQ) and global alignment and recovers their fine-grained details through cross-attention in the decoder for reconstruction. As for the second-stage generation, we introduce MergeAR, which performs KV Cache compression for efficient raster-order prediction. Extensive experiments on ImageNet verify that MergeVQ as an AR generative model achieves competitive performance in both visual representation learning and image generation tasks while maintaining favorable token efficiency and inference speed. The code and model will be available at https://apexgen-x.github.io/MergeVQ.
A Convex Formulation of Material Points and Rigid Bodies with GPU-Accelerated Async-Coupling for Interactive Simulation
Mar 06, 2025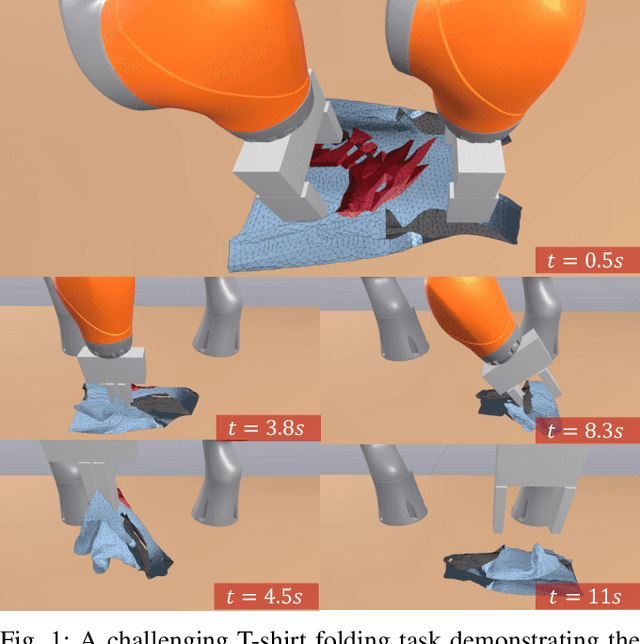
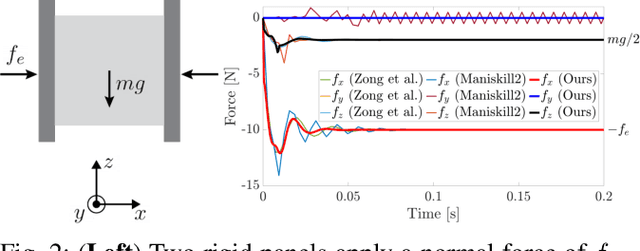
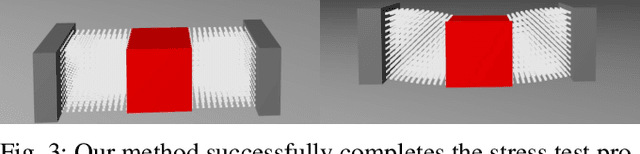
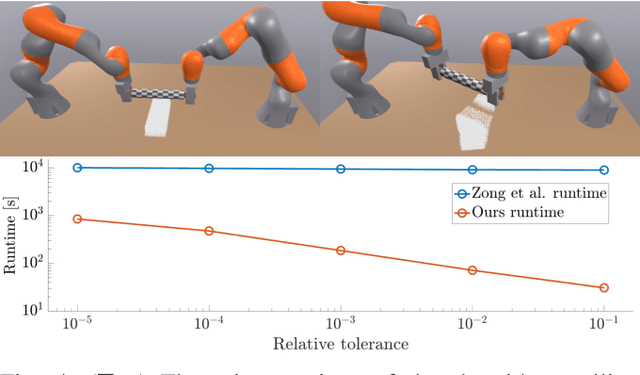
Abstract:We present a novel convex formulation that weakly couples the Material Point Method (MPM) with rigid body dynamics through frictional contact, optimized for efficient GPU parallelization. Our approach features an asynchronous time-splitting scheme to integrate MPM and rigid body dynamics under different time step sizes. We develop a globally convergent quasi-Newton solver tailored for massive parallelization, achieving up to 500x speedup over previous convex formulations without sacrificing stability. Our method enables interactive-rate simulations of robotic manipulation tasks with diverse deformable objects including granular materials and cloth, with strong convergence guarantees. We detail key implementation strategies to maximize performance and validate our approach through rigorous experiments, demonstrating superior speed, accuracy, and stability compared to state-of-the-art MPM simulators for robotics. We make our method available in the open-source robotics toolkit, Drake.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge