Jingbo Zhou
Unrewarded Exploration in Large Language Models Reveals Latent Learning from Psychology
Jan 30, 2026Abstract:Latent learning, classically theorized by Tolman, shows that biological agents (e.g., rats) can acquire internal representations of their environment without rewards, enabling rapid adaptation once rewards are introduced. In contrast, from a cognitive science perspective, reward learning remains overly dependent on external feedback, limiting flexibility and generalization. Although recent advances in the reasoning capabilities of large language models (LLMs), such as OpenAI-o1 and DeepSeek-R1, mark a significant breakthrough, these models still rely primarily on reward-centric reinforcement learning paradigms. Whether and how the well-established phenomenon of latent learning in psychology can inform or emerge within LLMs' training remains largely unexplored. In this work, we present novel findings from our experiments that LLMs also exhibit the latent learning dynamics. During an initial phase of unrewarded exploration, LLMs display modest performance improvements, as this phase allows LLMs to organize task-relevant knowledge without being constrained by reward-driven biases, and performance is further enhanced once rewards are introduced. LLMs post-trained under this two-stage exploration regime ultimately achieve higher competence than those post-trained with reward-based reinforcement learning throughout. Beyond these empirical observations, we also provide theoretical analyses for our experiments explaining why unrewarded exploration yields performance gains, offering a mechanistic account of these dynamics. Specifically, we conducted extensive experiments across multiple model families and diverse task domains to establish the existence of the latent learning dynamics in LLMs.
OmegaUse: Building a General-Purpose GUI Agent for Autonomous Task Execution
Jan 28, 2026Abstract:Graphical User Interface (GUI) agents show great potential for enabling foundation models to complete real-world tasks, revolutionizing human-computer interaction and improving human productivity. In this report, we present OmegaUse, a general-purpose GUI agent model for autonomous task execution on both mobile and desktop platforms, supporting computer-use and phone-use scenarios. Building an effective GUI agent model relies on two factors: (1) high-quality data and (2) effective training methods. To address these, we introduce a carefully engineered data-construction pipeline and a decoupled training paradigm. For data construction, we leverage rigorously curated open-source datasets and introduce a novel automated synthesis framework that integrates bottom-up autonomous exploration with top-down taxonomy-guided generation to create high-fidelity synthetic data. For training, to better leverage these data, we adopt a two-stage strategy: Supervised Fine-Tuning (SFT) to establish fundamental interaction syntax, followed by Group Relative Policy Optimization (GRPO) to improve spatial grounding and sequential planning. To balance computational efficiency with agentic reasoning capacity, OmegaUse is built on a Mixture-of-Experts (MoE) backbone. To evaluate cross-terminal capabilities in an offline setting, we introduce OS-Nav, a benchmark suite spanning multiple operating systems: ChiM-Nav, targeting Chinese Android mobile environments, and Ubu-Nav, focusing on routine desktop interactions on Ubuntu. Extensive experiments show that OmegaUse is highly competitive across established GUI benchmarks, achieving a state-of-the-art (SOTA) score of 96.3% on ScreenSpot-V2 and a leading 79.1% step success rate on AndroidControl. OmegaUse also performs strongly on OS-Nav, reaching 74.24% step success on ChiM-Nav and 55.9% average success on Ubu-Nav.
MergeDNA: Context-aware Genome Modeling with Dynamic Tokenization through Token Merging
Nov 17, 2025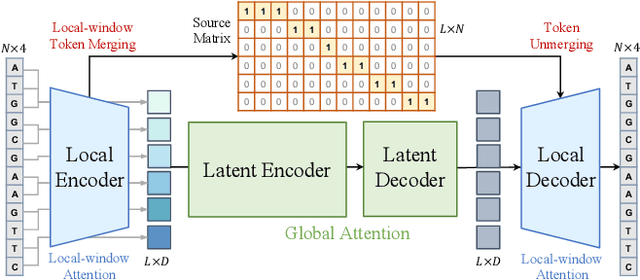
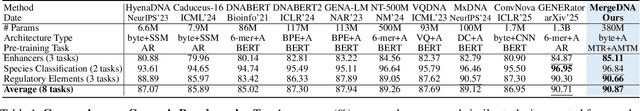
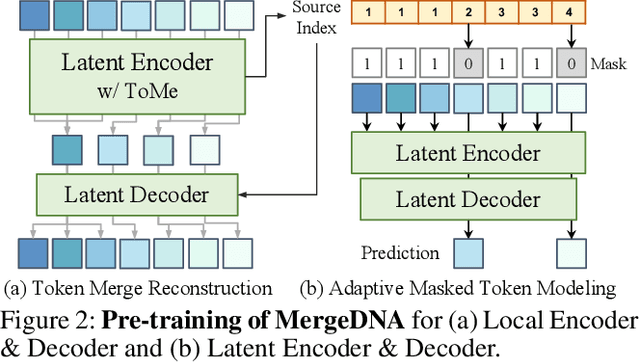
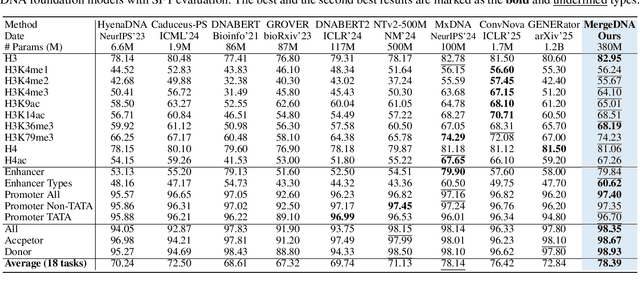
Abstract:Modeling genomic sequences faces two unsolved challenges: the information density varies widely across different regions, while there is no clearly defined minimum vocabulary unit. Relying on either four primitive bases or independently designed DNA tokenizers, existing approaches with naive masked language modeling pre-training often fail to adapt to the varying complexities of genomic sequences. Leveraging Token Merging techniques, this paper introduces a hierarchical architecture that jointly optimizes a dynamic genomic tokenizer and latent Transformers with context-aware pre-training tasks. As for network structures, the tokenization module automatically chunks adjacent bases into words by stacking multiple layers of the differentiable token merging blocks with local-window constraints, then a Latent Encoder captures the global context of these merged words by full-attention blocks. Symmetrically employing a Latent Decoder and a Local Decoder, MergeDNA learns with two pre-training tasks: Merged Token Reconstruction simultaneously trains the dynamic tokenization module and adaptively filters important tokens, while Adaptive Masked Token Modeling learns to predict these filtered tokens to capture informative contents. Extensive experiments show that MergeDNA achieves superior performance on three popular DNA benchmarks and several multi-omics tasks with fine-tuning or zero-shot evaluation, outperforming typical tokenization methods and large-scale DNA foundation models.
PRESCRIBE: Predicting Single-Cell Responses with Bayesian Estimation
Oct 09, 2025Abstract:In single-cell perturbation prediction, a central task is to forecast the effects of perturbing a gene unseen in the training data. The efficacy of such predictions depends on two factors: (1) the similarity of the target gene to those covered in the training data, which informs model (epistemic) uncertainty, and (2) the quality of the corresponding training data, which reflects data (aleatoric) uncertainty. Both factors are critical for determining the reliability of a prediction, particularly as gene perturbation is an inherently stochastic biochemical process. In this paper, we propose PRESCRIBE (PREdicting Single-Cell Response wIth Bayesian Estimation), a multivariate deep evidential regression framework designed to measure both sources of uncertainty jointly. Our analysis demonstrates that PRESCRIBE effectively estimates a confidence score for each prediction, which strongly correlates with its empirical accuracy. This capability enables the filtering of untrustworthy results, and in our experiments, it achieves steady accuracy improvements of over 3% compared to comparable baselines.
AAPO: Enhance the Reasoning Capabilities of LLMs with Advantage Momentum
May 20, 2025Abstract:Reinforcement learning (RL) has emerged as an effective approach for enhancing the reasoning capabilities of large language models (LLMs), especially in scenarios where supervised fine-tuning (SFT) falls short due to limited chain-of-thought (CoT) data. Among RL-based post-training methods, group relative advantage estimation, as exemplified by Group Relative Policy Optimization (GRPO), has attracted considerable attention for eliminating the dependency on the value model, thereby simplifying training compared to traditional approaches like Proximal Policy Optimization (PPO). However, we observe that exsiting group relative advantage estimation method still suffers from training inefficiencies, particularly when the estimated advantage approaches zero. To address this limitation, we propose Advantage-Augmented Policy Optimization (AAPO), a novel RL algorithm that optimizes the cross-entropy (CE) loss using advantages enhanced through a momentum-based estimation scheme. This approach effectively mitigates the inefficiencies associated with group relative advantage estimation. Experimental results on multiple mathematical reasoning benchmarks demonstrate the superior performance of AAPO.
GRAPE: Heterogeneous Graph Representation Learning for Genetic Perturbation with Coding and Non-Coding Biotype
May 06, 2025Abstract:Predicting genetic perturbations enables the identification of potentially crucial genes prior to wet-lab experiments, significantly improving overall experimental efficiency. Since genes are the foundation of cellular life, building gene regulatory networks (GRN) is essential to understand and predict the effects of genetic perturbations. However, current methods fail to fully leverage gene-related information, and solely rely on simple evaluation metrics to construct coarse-grained GRN. More importantly, they ignore functional differences between biotypes, limiting the ability to capture potential gene interactions. In this work, we leverage pre-trained large language model and DNA sequence model to extract features from gene descriptions and DNA sequence data, respectively, which serve as the initialization for gene representations. Additionally, we introduce gene biotype information for the first time in genetic perturbation, simulating the distinct roles of genes with different biotypes in regulating cellular processes, while capturing implicit gene relationships through graph structure learning (GSL). We propose GRAPE, a heterogeneous graph neural network (HGNN) that leverages gene representations initialized with features from descriptions and sequences, models the distinct roles of genes with different biotypes, and dynamically refines the GRN through GSL. The results on publicly available datasets show that our method achieves state-of-the-art performance.
ImageScope: Unifying Language-Guided Image Retrieval via Large Multimodal Model Collective Reasoning
Mar 13, 2025



Abstract:With the proliferation of images in online content, language-guided image retrieval (LGIR) has emerged as a research hotspot over the past decade, encompassing a variety of subtasks with diverse input forms. While the development of large multimodal models (LMMs) has significantly facilitated these tasks, existing approaches often address them in isolation, requiring the construction of separate systems for each task. This not only increases system complexity and maintenance costs, but also exacerbates challenges stemming from language ambiguity and complex image content, making it difficult for retrieval systems to provide accurate and reliable results. To this end, we propose ImageScope, a training-free, three-stage framework that leverages collective reasoning to unify LGIR tasks. The key insight behind the unification lies in the compositional nature of language, which transforms diverse LGIR tasks into a generalized text-to-image retrieval process, along with the reasoning of LMMs serving as a universal verification to refine the results. To be specific, in the first stage, we improve the robustness of the framework by synthesizing search intents across varying levels of semantic granularity using chain-of-thought (CoT) reasoning. In the second and third stages, we then reflect on retrieval results by verifying predicate propositions locally, and performing pairwise evaluations globally. Experiments conducted on six LGIR datasets demonstrate that ImageScope outperforms competitive baselines. Comprehensive evaluations and ablation studies further confirm the effectiveness of our design.
TimeFound: A Foundation Model for Time Series Forecasting
Mar 06, 2025



Abstract:We present TimeFound, an encoder-decoder transformer-based time series foundation model for out-of-the-box zero-shot forecasting. To handle time series data from various domains, TimeFound employs a multi-resolution patching strategy to capture complex temporal patterns at multiple scales. We pre-train our model with two sizes (200M and 710M parameters) on a large time-series corpus comprising both real-world and synthetic datasets. Over a collection of unseen datasets across diverse domains and forecasting horizons, our empirical evaluations suggest that TimeFound can achieve superior or competitive zero-shot forecasting performance, compared to state-of-the-art time series foundation models.
NodeReg: Mitigating the Imbalance and Distribution Shift Effects in Semi-Supervised Node Classification via Norm Consistency
Mar 05, 2025



Abstract:Aggregating information from neighboring nodes benefits graph neural networks (GNNs) in semi-supervised node classification tasks. Nevertheless, this mechanism also renders nodes susceptible to the influence of their neighbors. For instance, this will occur when the neighboring nodes are imbalanced or the neighboring nodes contain noise, which can even affect the GNN's ability to generalize out of distribution. We find that ensuring the consistency of the norm for node representations can significantly reduce the impact of these two issues on GNNs. To this end, we propose a regularized optimization method called NodeReg that enforces the consistency of node representation norms. This method is simple but effective and satisfies Lipschitz continuity, thus facilitating stable optimization and significantly improving semi-supervised node classification performance under the above two scenarios. To illustrate, in the imbalance scenario, when training a GCN with an imbalance ratio of 0.1, NodeReg outperforms the most competitive baselines by 1.4%-25.9% in F1 score across five public datasets. Similarly, in the distribution shift scenario, NodeReg outperforms the most competitive baseline by 1.4%-3.1% in accuracy.
CHAT: Beyond Contrastive Graph Transformer for Link Prediction in Heterogeneous Networks
Jan 06, 2025Abstract:Link prediction in heterogeneous networks is crucial for understanding the intricacies of network structures and forecasting their future developments. Traditional methodologies often face significant obstacles, including over-smoothing-wherein the excessive aggregation of node features leads to the loss of critical structural details-and a dependency on human-defined meta-paths, which necessitate extensive domain knowledge and can be inherently restrictive. These limitations hinder the effective prediction and analysis of complex heterogeneous networks. In response to these challenges, we propose the Contrastive Heterogeneous grAph Transformer (CHAT). CHAT introduces a novel sampling-based graph transformer technique that selectively retains nodes of interest, thereby obviating the need for predefined meta-paths. The method employs an innovative connection-aware transformer to encode node sequences and their interconnections with high fidelity, guided by a dual-faceted loss function specifically designed for heterogeneous network link prediction. Additionally, CHAT incorporates an ensemble link predictor that synthesizes multiple samplings to achieve enhanced prediction accuracy. We conducted comprehensive evaluations of CHAT using three distinct drug-target interaction (DTI) datasets. The empirical results underscore CHAT's superior performance, outperforming both general-task approaches and models specialized in DTI prediction. These findings substantiate the efficacy of CHAT in addressing the complex problem of link prediction in heterogeneous networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge