Yukun Zhou
GigaBrain-0.5M*: a VLA That Learns From World Model-Based Reinforcement Learning
Feb 12, 2026Abstract:Vision-language-action (VLA) models that directly predict multi-step action chunks from current observations face inherent limitations due to constrained scene understanding and weak future anticipation capabilities. In contrast, video world models pre-trained on web-scale video corpora exhibit robust spatiotemporal reasoning and accurate future prediction, making them a natural foundation for enhancing VLA learning. Therefore, we propose \textit{GigaBrain-0.5M*}, a VLA model trained via world model-based reinforcement learning. Built upon \textit{GigaBrain-0.5}, which is pre-trained on over 10,000 hours of robotic manipulation data, whose intermediate version currently ranks first on the international RoboChallenge benchmark. \textit{GigaBrain-0.5M*} further integrates world model-based reinforcement learning via \textit{RAMP} (Reinforcement leArning via world Model-conditioned Policy) to enable robust cross-task adaptation. Empirical results demonstrate that \textit{RAMP} achieves substantial performance gains over the RECAP baseline, yielding improvements of approximately 30\% on challenging tasks including \texttt{Laundry Folding}, \texttt{Box Packing}, and \texttt{Espresso Preparation}. Critically, \textit{GigaBrain-0.5M$^*$} exhibits reliable long-horizon execution, consistently accomplishing complex manipulation tasks without failure as validated by real-world deployment videos on our \href{https://gigabrain05m.github.io}{project page}.
ALPBench: A Benchmark for Attribution-level Long-term Personal Behavior Understanding
Feb 03, 2026Abstract:Recent advances in large language models have highlighted their potential for personalized recommendation, where accurately capturing user preferences remains a key challenge. Leveraging their strong reasoning and generalization capabilities, LLMs offer new opportunities for modeling long-term user behavior. To systematically evaluate this, we introduce ALPBench, a Benchmark for Attribution-level Long-term Personal Behavior Understanding. Unlike item-focused benchmarks, ALPBench predicts user-interested attribute combinations, enabling ground-truth evaluation even for newly introduced items. It models preferences from long-term historical behaviors rather than users' explicitly expressed requests, better reflecting enduring interests. User histories are represented as natural language sequences, allowing interpretable, reasoning-based personalization. ALPBench enables fine-grained evaluation of personalization by focusing on the prediction of attribute combinations task that remains highly challenging for current LLMs due to the need to capture complex interactions among multiple attributes and reason over long-term user behavior sequences.
oculomix: Hierarchical Sampling for Retinal-Based Systemic Disease Prediction
Jan 16, 2026Abstract:Oculomics - the concept of predicting systemic diseases, such as cardiovascular disease and dementia, through retinal imaging - has advanced rapidly due to the data efficiency of transformer-based foundation models like RETFound. Image-level mixed sample data augmentations, such as CutMix and MixUp, are frequently used for training transformers, yet these techniques perturb patient-specific attributes, such as medical comorbidity and clinical factors, since they only account for images and labels. To address this limitation, we propose a hierarchical sampling strategy, Oculomix, for mixed sample augmentations. Our method is based on two clinical priors. First (exam level), images acquired from the same patient at the same time point share the same attributes. Second (patient level), images acquired from the same patient at different time points have a soft temporal trend, as morbidity generally increases over time. Guided by these priors, our method constrains the mixing space to the patient and exam levels to better preserve patient-specific characteristics and leverages their hierarchical relationships. The proposed method is validated using ViT models on a five-year prediction of major adverse cardiovascular events (MACE) in a large ethnically diverse population (Alzeye). We show that Oculomix consistently outperforms image-level CutMix and MixUp by up to 3% in AUROC, demonstrating the necessity and value of the proposed method in oculomics.
Native Intelligence Emerges from Large-Scale Clinical Practice: A Retinal Foundation Model with Deployment Efficiency
Dec 16, 2025Abstract:Current retinal foundation models remain constrained by curated research datasets that lack authentic clinical context, and require extensive task-specific optimization for each application, limiting their deployment efficiency in low-resource settings. Here, we show that these barriers can be overcome by building clinical native intelligence directly from real-world medical practice. Our key insight is that large-scale telemedicine programs, where expert centers provide remote consultations across distributed facilities, represent a natural reservoir for learning clinical image interpretation. We present ReVision, a retinal foundation model that learns from the natural alignment between 485,980 color fundus photographs and their corresponding diagnostic reports, accumulated through a decade-long telemedicine program spanning 162 medical institutions across China. Through extensive evaluation across 27 ophthalmic benchmarks, we demonstrate that ReVison enables deployment efficiency with minimal local resources. Without any task-specific training, ReVision achieves zero-shot disease detection with an average AUROC of 0.946 across 12 public benchmarks and 0.952 on 3 independent clinical cohorts. When minimal adaptation is feasible, ReVision matches extensively fine-tuned alternatives while requiring orders of magnitude fewer trainable parameters and labeled examples. The learned representations also transfer effectively to new clinical sites, imaging domains, imaging modalities, and systemic health prediction tasks. In a prospective reader study with 33 ophthalmologists, ReVision's zero-shot assistance improved diagnostic accuracy by 14.8% across all experience levels. These results demonstrate that clinical native intelligence can be directly extracted from clinical archives without any further annotation to build medical AI systems suited to various low-resource settings.
Benchmarking Real-World Medical Image Classification with Noisy Labels: Challenges, Practice, and Outlook
Dec 10, 2025Abstract:Learning from noisy labels remains a major challenge in medical image analysis, where annotation demands expert knowledge and substantial inter-observer variability often leads to inconsistent or erroneous labels. Despite extensive research on learning with noisy labels (LNL), the robustness of existing methods in medical imaging has not been systematically assessed. To address this gap, we introduce LNMBench, a comprehensive benchmark for Label Noise in Medical imaging. LNMBench encompasses \textbf{10} representative methods evaluated across 7 datasets, 6 imaging modalities, and 3 noise patterns, establishing a unified and reproducible framework for robustness evaluation under realistic conditions. Comprehensive experiments reveal that the performance of existing LNL methods degrades substantially under high and real-world noise, highlighting the persistent challenges of class imbalance and domain variability in medical data. Motivated by these findings, we further propose a simple yet effective improvement to enhance model robustness under such conditions. The LNMBench codebase is publicly released to facilitate standardized evaluation, promote reproducible research, and provide practical insights for developing noise-resilient algorithms in both research and real-world medical applications.The codebase is publicly available on https://github.com/myyy777/LNMBench.
EMMA: Generalizing Real-World Robot Manipulation via Generative Visual Transfer
Sep 26, 2025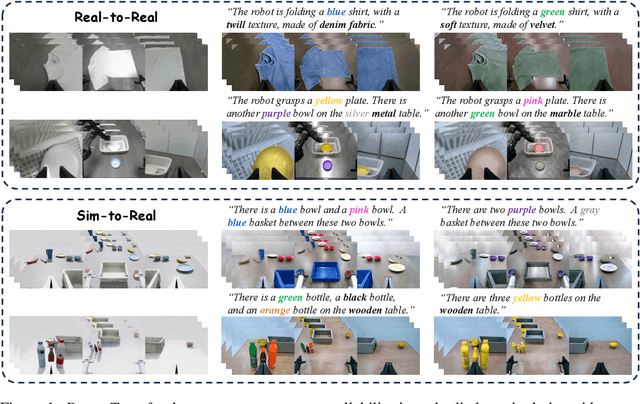
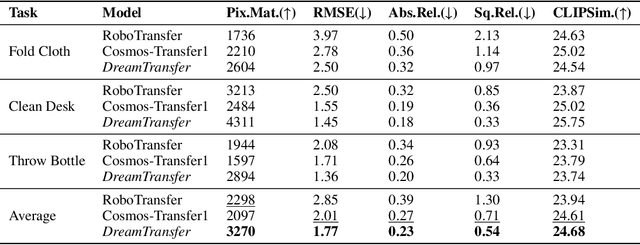
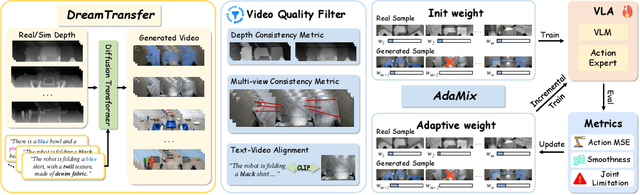
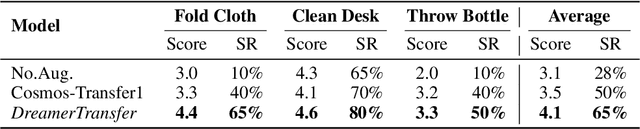
Abstract:Vision-language-action (VLA) models increasingly rely on diverse training data to achieve robust generalization. However, collecting large-scale real-world robot manipulation data across varied object appearances and environmental conditions remains prohibitively time-consuming and expensive. To overcome this bottleneck, we propose Embodied Manipulation Media Adaptation (EMMA), a VLA policy enhancement framework that integrates a generative data engine with an effective training pipeline. We introduce DreamTransfer, a diffusion Transformer-based framework for generating multi-view consistent, geometrically grounded embodied manipulation videos. DreamTransfer enables text-controlled visual editing of robot videos, transforming foreground, background, and lighting conditions without compromising 3D structure or geometrical plausibility. Furthermore, we explore hybrid training with real and generated data, and introduce AdaMix, a hard-sample-aware training strategy that dynamically reweights training batches to focus optimization on perceptually or kinematically challenging samples. Extensive experiments show that videos generated by DreamTransfer significantly outperform prior video generation methods in multi-view consistency, geometric fidelity, and text-conditioning accuracy. Crucially, VLAs trained with generated data enable robots to generalize to unseen object categories and novel visual domains using only demonstrations from a single appearance. In real-world robotic manipulation tasks with zero-shot visual domains, our approach achieves over a 200% relative performance gain compared to training on real data alone, and further improves by 13% with AdaMix, demonstrating its effectiveness in boosting policy generalization.
Generalist versus Specialist Vision Foundation Models for Ocular Disease and Oculomics
Sep 03, 2025Abstract:Medical foundation models, pre-trained with large-scale clinical data, demonstrate strong performance in diverse clinically relevant applications. RETFound, trained on nearly one million retinal images, exemplifies this approach in applications with retinal images. However, the emergence of increasingly powerful and multifold larger generalist foundation models such as DINOv2 and DINOv3 raises the question of whether domain-specific pre-training remains essential, and if so, what gap persists. To investigate this, we systematically evaluated the adaptability of DINOv2 and DINOv3 in retinal image applications, compared to two specialist RETFound models, RETFound-MAE and RETFound-DINOv2. We assessed performance on ocular disease detection and systemic disease prediction using two adaptation strategies: fine-tuning and linear probing. Data efficiency and adaptation efficiency were further analysed to characterise trade-offs between predictive performance and computational cost. Our results show that although scaling generalist models yields strong adaptability across diverse tasks, RETFound-DINOv2 consistently outperforms these generalist foundation models in ocular-disease detection and oculomics tasks, demonstrating stronger generalisability and data efficiency. These findings suggest that specialist retinal foundation models remain the most effective choice for clinical applications, while the narrowing gap with generalist foundation models suggests that continued data and model scaling can deliver domain-relevant gains and position them as strong foundations for future medical foundation models.
A computationally frugal open-source foundation model for thoracic disease detection in lung cancer screening programs
Jul 02, 2025Abstract:Low-dose computed tomography (LDCT) imaging employed in lung cancer screening (LCS) programs is increasing in uptake worldwide. LCS programs herald a generational opportunity to simultaneously detect cancer and non-cancer-related early-stage lung disease. Yet these efforts are hampered by a shortage of radiologists to interpret scans at scale. Here, we present TANGERINE, a computationally frugal, open-source vision foundation model for volumetric LDCT analysis. Designed for broad accessibility and rapid adaptation, TANGERINE can be fine-tuned off the shelf for a wide range of disease-specific tasks with limited computational resources and training data. Relative to models trained from scratch, TANGERINE demonstrates fast convergence during fine-tuning, thereby requiring significantly fewer GPU hours, and displays strong label efficiency, achieving comparable or superior performance with a fraction of fine-tuning data. Pretrained using self-supervised learning on over 98,000 thoracic LDCTs, including the UK's largest LCS initiative to date and 27 public datasets, TANGERINE achieves state-of-the-art performance across 14 disease classification tasks, including lung cancer and multiple respiratory diseases, while generalising robustly across diverse clinical centres. By extending a masked autoencoder framework to 3D imaging, TANGERINE offers a scalable solution for LDCT analysis, departing from recent closed, resource-intensive models by combining architectural simplicity, public availability, and modest computational requirements. Its accessible, open-source lightweight design lays the foundation for rapid integration into next-generation medical imaging tools that could transform LCS initiatives, allowing them to pivot from a singular focus on lung cancer detection to comprehensive respiratory disease management in high-risk populations.
HumanDreamer: Generating Controllable Human-Motion Videos via Decoupled Generation
Apr 01, 2025Abstract:Human-motion video generation has been a challenging task, primarily due to the difficulty inherent in learning human body movements. While some approaches have attempted to drive human-centric video generation explicitly through pose control, these methods typically rely on poses derived from existing videos, thereby lacking flexibility. To address this, we propose HumanDreamer, a decoupled human video generation framework that first generates diverse poses from text prompts and then leverages these poses to generate human-motion videos. Specifically, we propose MotionVid, the largest dataset for human-motion pose generation. Based on the dataset, we present MotionDiT, which is trained to generate structured human-motion poses from text prompts. Besides, a novel LAMA loss is introduced, which together contribute to a significant improvement in FID by 62.4%, along with respective enhancements in R-precision for top1, top2, and top3 by 41.8%, 26.3%, and 18.3%, thereby advancing both the Text-to-Pose control accuracy and FID metrics. Our experiments across various Pose-to-Video baselines demonstrate that the poses generated by our method can produce diverse and high-quality human-motion videos. Furthermore, our model can facilitate other downstream tasks, such as pose sequence prediction and 2D-3D motion lifting.
Is an Ultra Large Natural Image-Based Foundation Model Superior to a Retina-Specific Model for Detecting Ocular and Systemic Diseases?
Feb 10, 2025Abstract:The advent of foundation models (FMs) is transforming medical domain. In ophthalmology, RETFound, a retina-specific FM pre-trained sequentially on 1.4 million natural images and 1.6 million retinal images, has demonstrated high adaptability across clinical applications. Conversely, DINOv2, a general-purpose vision FM pre-trained on 142 million natural images, has shown promise in non-medical domains. However, its applicability to clinical tasks remains underexplored. To address this, we conducted head-to-head evaluations by fine-tuning RETFound and three DINOv2 models (large, base, small) for ocular disease detection and systemic disease prediction tasks, across eight standardized open-source ocular datasets, as well as the Moorfields AlzEye and the UK Biobank datasets. DINOv2-large model outperformed RETFound in detecting diabetic retinopathy (AUROC=0.850-0.952 vs 0.823-0.944, across three datasets, all P<=0.007) and multi-class eye diseases (AUROC=0.892 vs. 0.846, P<0.001). In glaucoma, DINOv2-base model outperformed RETFound (AUROC=0.958 vs 0.940, P<0.001). Conversely, RETFound achieved superior performance over all DINOv2 models in predicting heart failure, myocardial infarction, and ischaemic stroke (AUROC=0.732-0.796 vs 0.663-0.771, all P<0.001). These trends persisted even with 10% of the fine-tuning data. These findings showcase the distinct scenarios where general-purpose and domain-specific FMs excel, highlighting the importance of aligning FM selection with task-specific requirements to optimise clinical performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge