Joseph Jacob
A computationally frugal open-source foundation model for thoracic disease detection in lung cancer screening programs
Jul 02, 2025Abstract:Low-dose computed tomography (LDCT) imaging employed in lung cancer screening (LCS) programs is increasing in uptake worldwide. LCS programs herald a generational opportunity to simultaneously detect cancer and non-cancer-related early-stage lung disease. Yet these efforts are hampered by a shortage of radiologists to interpret scans at scale. Here, we present TANGERINE, a computationally frugal, open-source vision foundation model for volumetric LDCT analysis. Designed for broad accessibility and rapid adaptation, TANGERINE can be fine-tuned off the shelf for a wide range of disease-specific tasks with limited computational resources and training data. Relative to models trained from scratch, TANGERINE demonstrates fast convergence during fine-tuning, thereby requiring significantly fewer GPU hours, and displays strong label efficiency, achieving comparable or superior performance with a fraction of fine-tuning data. Pretrained using self-supervised learning on over 98,000 thoracic LDCTs, including the UK's largest LCS initiative to date and 27 public datasets, TANGERINE achieves state-of-the-art performance across 14 disease classification tasks, including lung cancer and multiple respiratory diseases, while generalising robustly across diverse clinical centres. By extending a masked autoencoder framework to 3D imaging, TANGERINE offers a scalable solution for LDCT analysis, departing from recent closed, resource-intensive models by combining architectural simplicity, public availability, and modest computational requirements. Its accessible, open-source lightweight design lays the foundation for rapid integration into next-generation medical imaging tools that could transform LCS initiatives, allowing them to pivot from a singular focus on lung cancer detection to comprehensive respiratory disease management in high-risk populations.
4D VQ-GAN: Synthesising Medical Scans at Any Time Point for Personalised Disease Progression Modelling of Idiopathic Pulmonary Fibrosis
Feb 08, 2025Abstract:Understanding the progression trajectories of diseases is crucial for early diagnosis and effective treatment planning. This is especially vital for life-threatening conditions such as Idiopathic Pulmonary Fibrosis (IPF), a chronic, progressive lung disease with a prognosis comparable to many cancers. Computed tomography (CT) imaging has been established as a reliable diagnostic tool for IPF. Accurately predicting future CT scans of early-stage IPF patients can aid in developing better treatment strategies, thereby improving survival outcomes. In this paper, we propose 4D Vector Quantised Generative Adversarial Networks (4D-VQ-GAN), a model capable of generating realistic CT volumes of IPF patients at any time point. The model is trained using a two-stage approach. In the first stage, a 3D-VQ-GAN is trained to reconstruct CT volumes. In the second stage, a Neural Ordinary Differential Equation (ODE) based temporal model is trained to capture the temporal dynamics of the quantised embeddings generated by the encoder in the first stage. We evaluate different configurations of our model for generating longitudinal CT scans and compare the results against ground truth data, both quantitatively and qualitatively. For validation, we conduct survival analysis using imaging biomarkers derived from generated CT scans and achieve a C-index comparable to that of biomarkers derived from the real CT scans. The survival analysis results demonstrate the potential clinical utility inherent to generated longitudinal CT scans, showing that they can reliably predict survival outcomes.
MRI Parameter Mapping via Gaussian Mixture VAE: Breaking the Assumption of Independent Pixels
Nov 16, 2024



Abstract:We introduce and demonstrate a new paradigm for quantitative parameter mapping in MRI. Parameter mapping techniques, such as diffusion MRI and quantitative MRI, have the potential to robustly and repeatably measure biologically-relevant tissue maps that strongly relate to underlying microstructure. Quantitative maps are calculated by fitting a model to multiple images, e.g. with least-squares or machine learning. However, the overwhelming majority of model fitting techniques assume that each voxel is independent, ignoring any co-dependencies in the data. This makes model fitting sensitive to voxelwise measurement noise, hampering reliability and repeatability. We propose a self-supervised deep variational approach that breaks the assumption of independent pixels, leveraging redundancies in the data to effectively perform data-driven regularisation of quantitative maps. We demonstrate that our approach outperforms current model fitting techniques in dMRI simulations and real data. Especially with a Gaussian mixture prior, our model enables sharper quantitative maps, revealing finer anatomical details that are not presented in the baselines. Our approach can hence support the clinical adoption of parameter mapping methods such as dMRI and qMRI.
Challenges for Responsible AI Design and Workflow Integration in Healthcare: A Case Study of Automatic Feeding Tube Qualification in Radiology
May 08, 2024



Abstract:Nasogastric tubes (NGTs) are feeding tubes that are inserted through the nose into the stomach to deliver nutrition or medication. If not placed correctly, they can cause serious harm, even death to patients. Recent AI developments demonstrate the feasibility of robustly detecting NGT placement from Chest X-ray images to reduce risks of sub-optimally or critically placed NGTs being missed or delayed in their detection, but gaps remain in clinical practice integration. In this study, we present a human-centered approach to the problem and describe insights derived following contextual inquiry and in-depth interviews with 15 clinical stakeholders. The interviews helped understand challenges in existing workflows, and how best to align technical capabilities with user needs and expectations. We discovered the trade-offs and complexities that need consideration when choosing suitable workflow stages, target users, and design configurations for different AI proposals. We explored how to balance AI benefits and risks for healthcare staff and patients within broader organizational and medical-legal constraints. We also identified data issues related to edge cases and data biases that affect model training and evaluation; how data documentation practices influence data preparation and labelling; and how to measure relevant AI outcomes reliably in future evaluations. We discuss how our work informs design and development of AI applications that are clinically useful, ethical, and acceptable in real-world healthcare services.
CenTime: Event-Conditional Modelling of Censoring in Survival Analysis
Sep 15, 2023Abstract:Survival analysis is a valuable tool for estimating the time until specific events, such as death or cancer recurrence, based on baseline observations. This is particularly useful in healthcare to prognostically predict clinically important events based on patient data. However, existing approaches often have limitations; some focus only on ranking patients by survivability, neglecting to estimate the actual event time, while others treat the problem as a classification task, ignoring the inherent time-ordered structure of the events. Furthermore, the effective utilization of censored samples - training data points where the exact event time is unknown - is essential for improving the predictive accuracy of the model. In this paper, we introduce CenTime, a novel approach to survival analysis that directly estimates the time to event. Our method features an innovative event-conditional censoring mechanism that performs robustly even when uncensored data is scarce. We demonstrate that our approach forms a consistent estimator for the event model parameters, even in the absence of uncensored data. Furthermore, CenTime is easily integrated with deep learning models with no restrictions on batch size or the number of uncensored samples. We compare our approach with standard survival analysis methods, including the Cox proportional-hazard model and DeepHit. Our results indicate that CenTime offers state-of-the-art performance in predicting time-to-death while maintaining comparable ranking performance. Our implementation is publicly available at https://github.com/ahmedhshahin/CenTime.
A 3D deep learning classifier and its explainability when assessing coronary artery disease
Jul 29, 2023Abstract:Early detection and diagnosis of coronary artery disease (CAD) could save lives and reduce healthcare costs. In this study, we propose a 3D Resnet-50 deep learning model to directly classify normal subjects and CAD patients on computed tomography coronary angiography images. Our proposed method outperforms a 2D Resnet-50 model by 23.65%. Explainability is also provided by using a Grad-GAM. Furthermore, we link the 3D CAD classification to a 2D two-class semantic segmentation for improved explainability and accurate abnormality localisation.
A data-centric deep learning approach to airway segmentation
Jul 29, 2023Abstract:The morphology and distribution of airway tree abnormalities enables diagnosis and disease characterisation across a variety of chronic respiratory conditions. In this regard, airway segmentation plays a critical role in the production of the outline of the entire airway tree to enable estimation of disease extent and severity. In this study, we propose a data-centric deep learning technique to segment the airway tree. The proposed technique utilises interpolation and image split to improve data usefulness and quality. Then, an ensemble learning strategy is implemented to aggregate the segmented airway trees at different scales. In terms of segmentation performance (dice similarity coefficient), our method outperforms the baseline model by 2.5% on average when a combined loss is used. Further, our proposed technique has a low GPU usage and high flexibility enabling it to be deployed on any 2D deep learning model.
Expectation Maximization Pseudo Labelling for Segmentation with Limited Annotations
May 02, 2023Abstract:We study pseudo labelling and its generalisation for semi-supervised segmentation of medical images. Pseudo labelling has achieved great empirical successes in semi-supervised learning, by utilising raw inferences on unlabelled data as pseudo labels for self-training. In our paper, we build a connection between pseudo labelling and the Expectation Maximization algorithm which partially explains its empirical successes. We thereby realise that the original pseudo labelling is an empirical estimation of its underlying full formulation. Following this insight, we demonstrate the full generalisation of pseudo labels under Bayes' principle, called Bayesian Pseudo Labels. We then provide a variational approach to learn to approximate Bayesian Pseudo Labels, by learning a threshold to select good quality pseudo labels. In the rest of the paper, we demonstrate the applications of Pseudo Labelling and its generalisation Bayesian Psuedo Labelling in semi-supervised segmentation of medical images on: 1) 3D binary segmentation of lung vessels from CT volumes; 2) 2D multi class segmentation of brain tumours from MRI volumes; 3) 3D binary segmentation of brain tumours from MRI volumes. We also show that pseudo labels can enhance the robustness of the learnt representations.
A hybrid CNN-RNN approach for survival analysis in a Lung Cancer Screening study
Mar 19, 2023



Abstract:In this study, we present a hybrid CNN-RNN approach to investigate long-term survival of subjects in a lung cancer screening study. Subjects who died of cardiovascular and respiratory causes were identified whereby the CNN model was used to capture imaging features in the CT scans and the RNN model was used to investigate time series and thus global information. The models were trained on subjects who underwent cardiovascular and respiratory deaths and a control cohort matched to participant age, gender, and smoking history. The combined model can achieve an AUC of 0.76 which outperforms humans at cardiovascular mortality prediction. The corresponding F1 and Matthews Correlation Coefficient are 0.63 and 0.42 respectively. The generalisability of the model is further validated on an 'external' cohort. The same models were applied to survival analysis with the Cox Proportional Hazard model. It was demonstrated that incorporating the follow-up history can lead to improvement in survival prediction. The Cox neural network can achieve an IPCW C-index of 0.75 on the internal dataset and 0.69 on an external dataset. Delineating imaging features associated with long-term survival can help focus preventative interventions appropriately, particularly for under-recognised pathologies thereby potentially reducing patient morbidity.
Airway measurement by refinement of synthetic images improves mortality prediction in idiopathic pulmonary fibrosis
Aug 30, 2022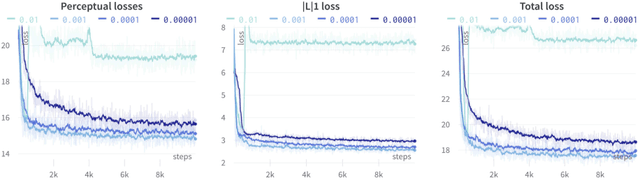
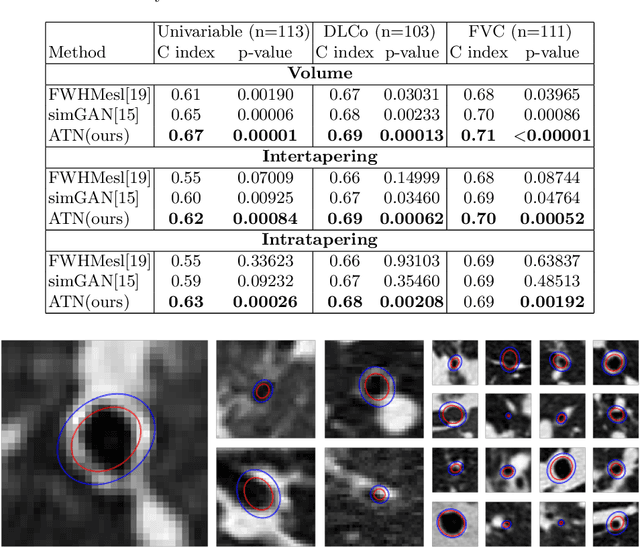
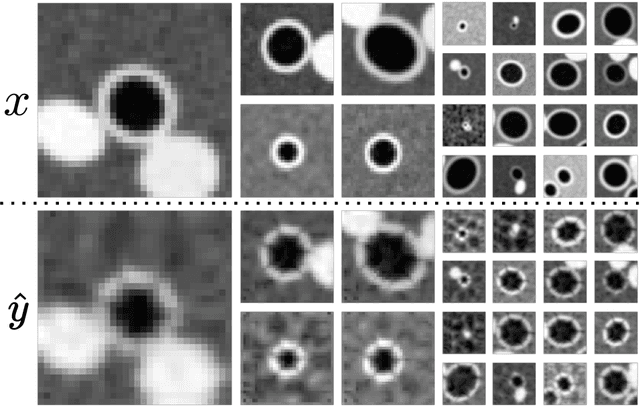
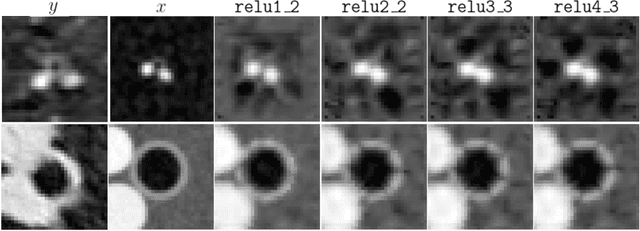
Abstract:Several chronic lung diseases, like idiopathic pulmonary fibrosis (IPF) are characterised by abnormal dilatation of the airways. Quantification of airway features on computed tomography (CT) can help characterise disease progression. Physics based airway measurement algorithms have been developed, but have met with limited success in part due to the sheer diversity of airway morphology seen in clinical practice. Supervised learning methods are also not feasible due to the high cost of obtaining precise airway annotations. We propose synthesising airways by style transfer using perceptual losses to train our model, Airway Transfer Network (ATN). We compare our ATN model with a state-of-the-art GAN-based network (simGAN) using a) qualitative assessment; b) assessment of the ability of ATN and simGAN based CT airway metrics to predict mortality in a population of 113 patients with IPF. ATN was shown to be quicker and easier to train than simGAN. ATN-based airway measurements were also found to be consistently stronger predictors of mortality than simGAN-derived airway metrics on IPF CTs. Airway synthesis by a transformation network that refines synthetic data using perceptual losses is a realistic alternative to GAN-based methods for clinical CT analyses of idiopathic pulmonary fibrosis. Our source code can be found at https://github.com/ashkanpakzad/ATN that is compatible with the existing open-source airway analysis framework, AirQuant.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge