Jieneng Chen
3D-TransUNet for Brain Metastases Segmentation in the BraTS2023 Challenge
Mar 23, 2024



Abstract:Segmenting brain tumors is complex due to their diverse appearances and scales. Brain metastases, the most common type of brain tumor, are a frequent complication of cancer. Therefore, an effective segmentation model for brain metastases must adeptly capture local intricacies to delineate small tumor regions while also integrating global context to understand broader scan features. The TransUNet model, which combines Transformer self-attention with U-Net's localized information, emerges as a promising solution for this task. In this report, we address brain metastases segmentation by training the 3D-TransUNet model on the Brain Tumor Segmentation (BraTS-METS) 2023 challenge dataset. Specifically, we explored two architectural configurations: the Encoder-only 3D-TransUNet, employing Transformers solely in the encoder, and the Decoder-only 3D-TransUNet, utilizing Transformers exclusively in the decoder. For Encoder-only 3D-TransUNet, we note that Masked-Autoencoder pre-training is required for a better initialization of the Transformer Encoder and thus accelerates the training process. We identify that the Decoder-only 3D-TransUNet model should offer enhanced efficacy in the segmentation of brain metastases, as indicated by our 5-fold cross-validation on the training set. However, our use of the Encoder-only 3D-TransUNet model already yield notable results, with an average lesion-wise Dice score of 59.8\% on the test set, securing second place in the BraTS-METS 2023 challenge.
Prompt-Based Exemplar Super-Compression and Regeneration for Class-Incremental Learning
Nov 30, 2023Abstract:Replay-based methods in class-incremental learning (CIL) have attained remarkable success, as replaying the exemplars of old classes can significantly mitigate catastrophic forgetting. Despite their effectiveness, the inherent memory restrictions of CIL result in saving a limited number of exemplars with poor diversity, leading to data imbalance and overfitting issues. In this paper, we introduce a novel exemplar super-compression and regeneration method, ESCORT, which substantially increases the quantity and enhances the diversity of exemplars. Rather than storing past images, we compress images into visual and textual prompts, e.g., edge maps and class tags, and save the prompts instead, reducing the memory usage of each exemplar to 1/24 of the original size. In subsequent learning phases, diverse high-resolution exemplars are generated from the prompts by a pre-trained diffusion model, e.g., ControlNet. To minimize the domain gap between generated exemplars and real images, we propose partial compression and diffusion-based data augmentation, allowing us to utilize an off-the-shelf diffusion model without fine-tuning it on the target dataset. Therefore, the same diffusion model can be downloaded whenever it is needed, incurring no memory consumption. Comprehensive experiments demonstrate that our method significantly improves model performance across multiple CIL benchmarks, e.g., 5.0 percentage points higher than the previous state-of-the-art on 10-phase Caltech-256 dataset.
3D TransUNet: Advancing Medical Image Segmentation through Vision Transformers
Oct 11, 2023Abstract:Medical image segmentation plays a crucial role in advancing healthcare systems for disease diagnosis and treatment planning. The u-shaped architecture, popularly known as U-Net, has proven highly successful for various medical image segmentation tasks. However, U-Net's convolution-based operations inherently limit its ability to model long-range dependencies effectively. To address these limitations, researchers have turned to Transformers, renowned for their global self-attention mechanisms, as alternative architectures. One popular network is our previous TransUNet, which leverages Transformers' self-attention to complement U-Net's localized information with the global context. In this paper, we extend the 2D TransUNet architecture to a 3D network by building upon the state-of-the-art nnU-Net architecture, and fully exploring Transformers' potential in both the encoder and decoder design. We introduce two key components: 1) A Transformer encoder that tokenizes image patches from a convolution neural network (CNN) feature map, enabling the extraction of global contexts, and 2) A Transformer decoder that adaptively refines candidate regions by utilizing cross-attention between candidate proposals and U-Net features. Our investigations reveal that different medical tasks benefit from distinct architectural designs. The Transformer encoder excels in multi-organ segmentation, where the relationship among organs is crucial. On the other hand, the Transformer decoder proves more beneficial for dealing with small and challenging segmented targets such as tumor segmentation. Extensive experiments showcase the significant potential of integrating a Transformer-based encoder and decoder into the u-shaped medical image segmentation architecture. TransUNet outperforms competitors in various medical applications.
Compositor: Bottom-up Clustering and Compositing for Robust Part and Object Segmentation
Jun 15, 2023Abstract:In this work, we present a robust approach for joint part and object segmentation. Specifically, we reformulate object and part segmentation as an optimization problem and build a hierarchical feature representation including pixel, part, and object-level embeddings to solve it in a bottom-up clustering manner. Pixels are grouped into several clusters where the part-level embeddings serve as cluster centers. Afterwards, object masks are obtained by compositing the part proposals. This bottom-up interaction is shown to be effective in integrating information from lower semantic levels to higher semantic levels. Based on that, our novel approach Compositor produces part and object segmentation masks simultaneously while improving the mask quality. Compositor achieves state-of-the-art performance on PartImageNet and Pascal-Part by outperforming previous methods by around 0.9% and 1.3% on PartImageNet, 0.4% and 1.7% on Pascal-Part in terms of part and object mIoU and demonstrates better robustness against occlusion by around 4.4% and 7.1% on part and object respectively. Code will be available at https://github.com/TACJu/Compositor.
Label-Free Liver Tumor Segmentation
Mar 27, 2023



Abstract:We demonstrate that AI models can accurately segment liver tumors without the need for manual annotation by using synthetic tumors in CT scans. Our synthetic tumors have two intriguing advantages: (I) realistic in shape and texture, which even medical professionals can confuse with real tumors; (II) effective for training AI models, which can perform liver tumor segmentation similarly to the model trained on real tumors -- this result is exciting because no existing work, using synthetic tumors only, has thus far reached a similar or even close performance to real tumors. This result also implies that manual efforts for annotating tumors voxel by voxel (which took years to create) can be significantly reduced in the future. Moreover, our synthetic tumors can automatically generate many examples of small (or even tiny) synthetic tumors and have the potential to improve the success rate of detecting small liver tumors, which is critical for detecting the early stages of cancer. In addition to enriching the training data, our synthesizing strategy also enables us to rigorously assess the AI robustness.
Towards a Single Unified Model for Effective Detection, Segmentation, and Diagnosis of Eight Major Cancers Using a Large Collection of CT Scans
Jan 28, 2023



Abstract:Human readers or radiologists routinely perform full-body multi-organ multi-disease detection and diagnosis in clinical practice, while most medical AI systems are built to focus on single organs with a narrow list of a few diseases. This might severely limit AI's clinical adoption. A certain number of AI models need to be assembled non-trivially to match the diagnostic process of a human reading a CT scan. In this paper, we construct a Unified Tumor Transformer (UniT) model to detect (tumor existence and location) and diagnose (tumor characteristics) eight major cancer-prevalent organs in CT scans. UniT is a query-based Mask Transformer model with the output of multi-organ and multi-tumor semantic segmentation. We decouple the object queries into organ queries, detection queries and diagnosis queries, and further establish hierarchical relationships among the three groups. This clinically-inspired architecture effectively assists inter- and intra-organ representation learning of tumors and facilitates the resolution of these complex, anatomically related multi-organ cancer image reading tasks. UniT is trained end-to-end using a curated large-scale CT images of 10,042 patients including eight major types of cancers and occurring non-cancer tumors (all are pathology-confirmed with 3D tumor masks annotated by radiologists). On the test set of 631 patients, UniT has demonstrated strong performance under a set of clinically relevant evaluation metrics, substantially outperforming both multi-organ segmentation methods and an assembly of eight single-organ expert models in tumor detection, segmentation, and diagnosis. Such a unified multi-cancer image reading model (UniT) can significantly reduce the number of false positives produced by combined multi-system models. This moves one step closer towards a universal high-performance cancer screening tool.
MT-TransUNet: Mediating Multi-Task Tokens in Transformers for Skin Lesion Segmentation and Classification
Dec 03, 2021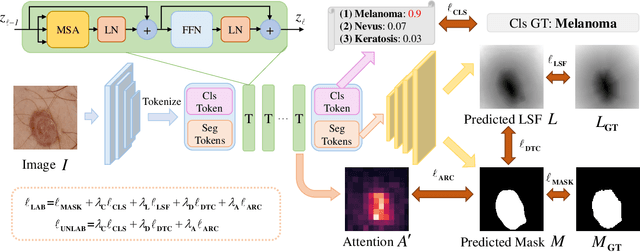
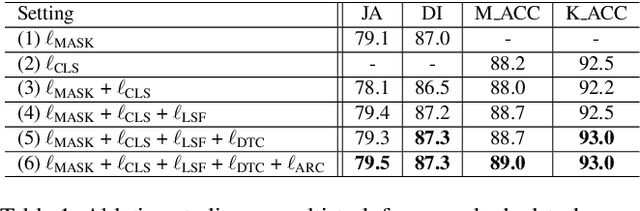
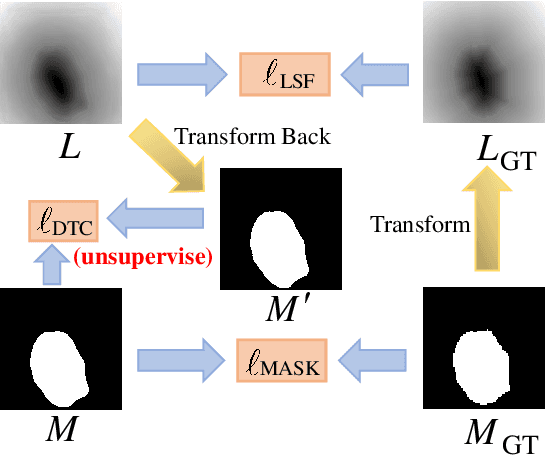
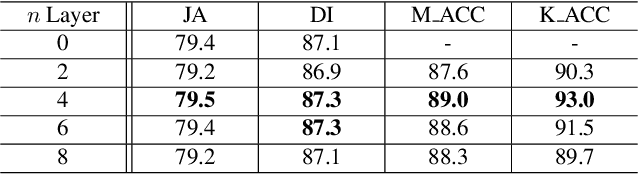
Abstract:Recent advances in automated skin cancer diagnosis have yielded performance on par with board-certified dermatologists. However, these approaches formulated skin cancer diagnosis as a simple classification task, dismissing the potential benefit from lesion segmentation. We argue that an accurate lesion segmentation can supplement the classification task with additive lesion information, such as asymmetry, border, intensity, and physical size; in turn, a faithful lesion classification can support the segmentation task with discriminant lesion features. To this end, this paper proposes a new multi-task framework, named MT-TransUNet, which is capable of segmenting and classifying skin lesions collaboratively by mediating multi-task tokens in Transformers. Furthermore, we have introduced dual-task and attended region consistency losses to take advantage of those images without pixel-level annotation, ensuring the model's robustness when it encounters the same image with an account of augmentation. Our MT-TransUNet exceeds the previous state of the art for lesion segmentation and classification tasks in ISIC-2017 and PH2; more importantly, it preserves compelling computational efficiency regarding model parameters (48M~vs.~130M) and inference speed (0.17s~vs.~2.02s per image). Code will be available at https://github.com/JingyeChen/MT-TransUNet.
SCPM-Net: An Anchor-free 3D Lung Nodule Detection Network using Sphere Representation and Center Points Matching
Apr 12, 2021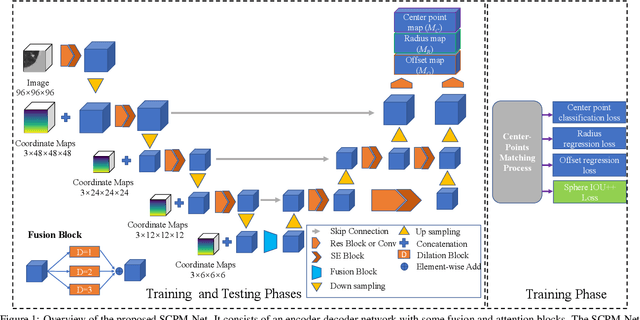
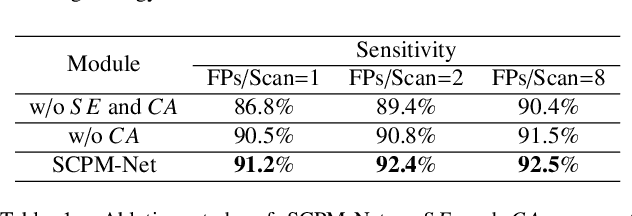
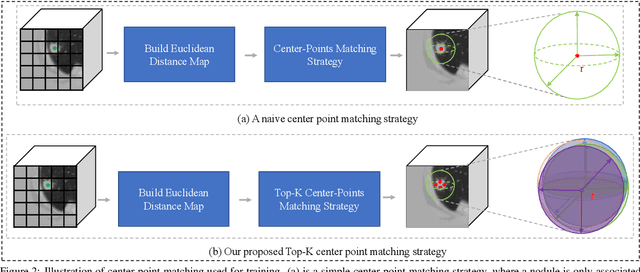
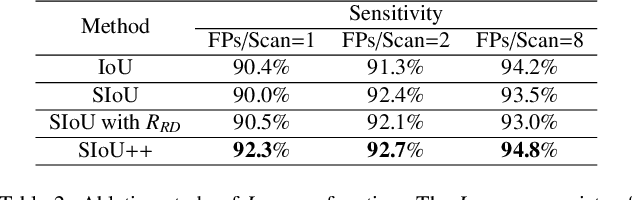
Abstract:Automatic and accurate lung nodule detection from 3D Computed Tomography scans plays a vital role in efficient lung cancer screening. Despite the state-of-the-art performance obtained by recent anchor-based detectors using Convolutional Neural Networks, they require predetermined anchor parameters such as the size, number, and aspect ratio of anchors, and have limited robustness when dealing with lung nodules with a massive variety of sizes. We propose a 3D sphere representation-based center-points matching detection network (SCPM-Net) that is anchor-free and automatically predicts the position, radius, and offset of nodules without the manual design of nodule/anchor parameters. The SCPM-Net consists of two novel pillars: sphere representation and center points matching. To mimic the nodule annotation in clinical practice, we replace the conventional bounding box with the newly proposed bounding sphere. A compatible sphere-based intersection over-union loss function is introduced to train the lung nodule detection network stably and efficiently.We empower the network anchor-free by designing a positive center-points selection and matching (CPM) process, which naturally discards pre-determined anchor boxes. An online hard example mining and re-focal loss subsequently enable the CPM process more robust, resulting in more accurate point assignment and the mitigation of class imbalance. In addition, to better capture spatial information and 3D context for the detection, we propose to fuse multi-level spatial coordinate maps with the feature extractor and combine them with 3D squeeze-and-excitation attention modules. Experimental results on the LUNA16 dataset showed that our proposed SCPM-Net framework achieves superior performance compared with existing used anchor-based and anchor-free methods for lung nodule detection.
Sequential Learning on Liver Tumor Boundary Semantics and Prognostic Biomarker Mining
Mar 09, 2021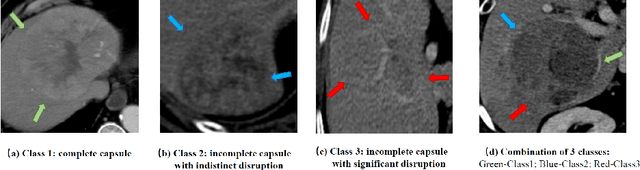
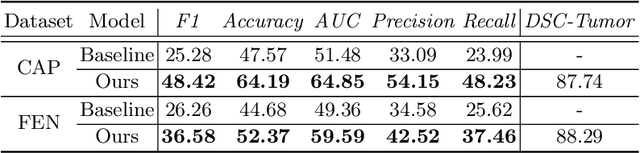
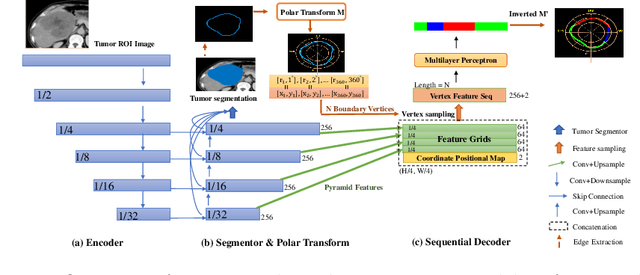
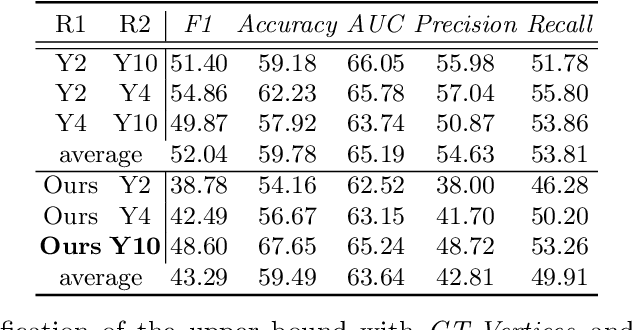
Abstract:The boundary of tumors (hepatocellular carcinoma, or HCC) contains rich semantics: capsular invasion, visibility, smoothness, folding and protuberance, etc. Capsular invasion on tumor boundary has proven to be clinically correlated with the prognostic indicator, microvascular invasion (MVI). Investigating tumor boundary semantics has tremendous clinical values. In this paper, we propose the first and novel computational framework that disentangles the task into two components: spatial vertex localization and sequential semantic classification. (1) A HCC tumor segmentor is built for tumor mask boundary extraction, followed by polar transform representing the boundary with radius and angle. Vertex generator is used to produce fixed-length boundary vertices where vertex features are sampled on the corresponding spatial locations. (2) The sampled deep vertex features with positional embedding are mapped into a sequential space and decoded by a multilayer perceptron (MLP) for semantic classification. Extensive experiments on tumor capsule semantics demonstrate the effectiveness of our framework. Mining the correlation between the boundary semantics and MVI status proves the feasibility to integrate this boundary semantics as a valid HCC prognostic biomarker.
SpecTr: Spectral Transformer for Hyperspectral Pathology Image Segmentation
Mar 05, 2021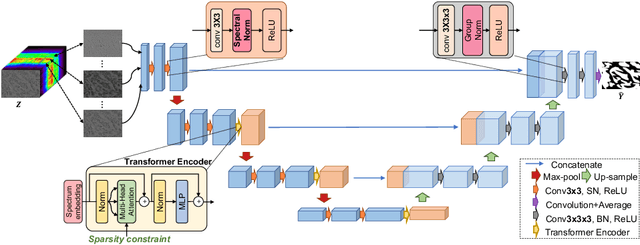
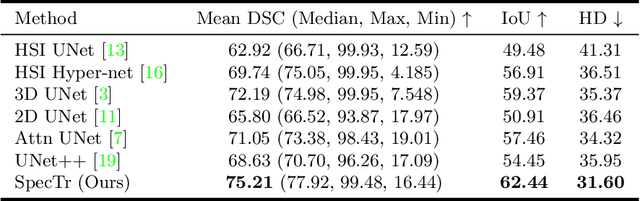
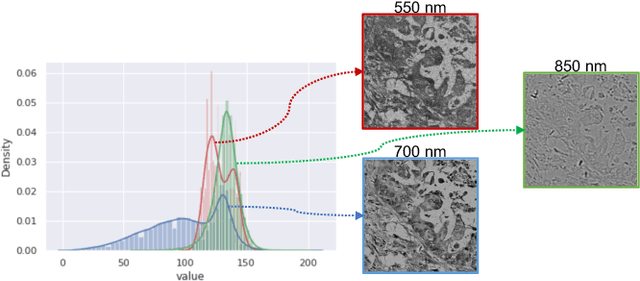
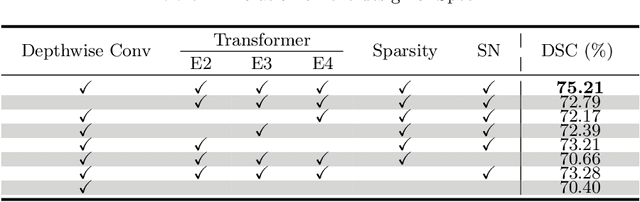
Abstract:Hyperspectral imaging (HSI) unlocks the huge potential to a wide variety of applications relied on high-precision pathology image segmentation, such as computational pathology and precision medicine. Since hyperspectral pathology images benefit from the rich and detailed spectral information even beyond the visible spectrum, the key to achieve high-precision hyperspectral pathology image segmentation is to felicitously model the context along high-dimensional spectral bands. Inspired by the strong context modeling ability of transformers, we hereby, for the first time, formulate the contextual feature learning across spectral bands for hyperspectral pathology image segmentation as a sequence-to-sequence prediction procedure by transformers. To assist spectral context learning procedure, we introduce two important strategies: (1) a sparsity scheme enforces the learned contextual relationship to be sparse, so as to eliminates the distraction from the redundant bands; (2) a spectral normalization, a separate group normalization for each spectral band, mitigates the nuisance caused by heterogeneous underlying distributions of bands. We name our method Spectral Transformer (SpecTr), which enjoys two benefits: (1) it has a strong ability to model long-range dependency among spectral bands, and (2) it jointly explores the spatial-spectral features of HSI. Experiments show that SpecTr outperforms other competing methods in a hyperspectral pathology image segmentation benchmark without the need of pre-training. Code is available at https://github.com/hfut-xc-yun/SpecTr.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge