Guotai Wang
University of Electronic Science and Technology of China, Chengdu, China, ShangAI Laboratory, Shanghai, China
A3-TTA: Adaptive Anchor Alignment Test-Time Adaptation for Image Segmentation
Feb 03, 2026Abstract:Test-Time Adaptation (TTA) offers a practical solution for deploying image segmentation models under domain shift without accessing source data or retraining. Among existing TTA strategies, pseudo-label-based methods have shown promising performance. However, they often rely on perturbation-ensemble heuristics (e.g., dropout sampling, test-time augmentation, Gaussian noise), which lack distributional grounding and yield unstable training signals. This can trigger error accumulation and catastrophic forgetting during adaptation. To address this, we propose \textbf{A3-TTA}, a TTA framework that constructs reliable pseudo-labels through anchor-guided supervision. Specifically, we identify well-predicted target domain images using a class compact density metric, under the assumption that confident predictions imply distributional proximity to the source domain. These anchors serve as stable references to guide pseudo-label generation, which is further regularized via semantic consistency and boundary-aware entropy minimization. Additionally, we introduce a self-adaptive exponential moving average strategy to mitigate label noise and stabilize model update during adaptation. Evaluated on both multi-domain medical images (heart structure and prostate segmentation) and natural images, A3-TTA significantly improves average Dice scores by 10.40 to 17.68 percentage points compared to the source model, outperforming several state-of-the-art TTA methods under different segmentation model architectures. A3-TTA also excels in continual TTA, maintaining high performance across sequential target domains with strong anti-forgetting ability. The code will be made publicly available at https://github.com/HiLab-git/A3-TTA.
SegRap2025: A Benchmark of Gross Tumor Volume and Lymph Node Clinical Target Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Jan 28, 2026Abstract:Accurate delineation of Gross Tumor Volume (GTV), Lymph Node Clinical Target Volume (LN CTV), and Organ-at-Risk (OAR) from Computed Tomography (CT) scans is essential for precise radiotherapy planning in Nasopharyngeal Carcinoma (NPC). Building upon SegRap2023, which focused on OAR and GTV segmentation using single-center paired non-contrast CT (ncCT) and contrast-enhanced CT (ceCT) scans, the SegRap2025 challenge aims to enhance the generalizability and robustness of segmentation models across imaging centers and modalities. SegRap2025 comprises two tasks: Task01 addresses GTV segmentation using paired CT from the SegRap2023 dataset, with an additional external testing set to evaluate cross-center generalization, and Task02 focuses on LN CTV segmentation using multi-center training data and an unseen external testing set, where each case contains paired CT scans or a single modality, emphasizing both cross-center and cross-modality robustness. This paper presents the challenge setup and provides a comprehensive analysis of the solutions submitted by ten participating teams. For GTV segmentation task, the top-performing models achieved average Dice Similarity Coefficient (DSC) of 74.61% and 56.79% on the internal and external testing cohorts, respectively. For LN CTV segmentation task, the highest average DSC values reached 60.24%, 60.50%, and 57.23% on paired CT, ceCT-only, and ncCT-only subsets, respectively. SegRap2025 establishes a large-scale multi-center, multi-modality benchmark for evaluating the generalization and robustness in radiotherapy target segmentation, providing valuable insights toward clinically applicable automated radiotherapy planning systems. The benchmark is available at: https://hilab-git.github.io/SegRap2025_Challenge.
InvCoSS: Inversion-driven Continual Self-supervised Learning in Medical Multi-modal Image Pre-training
Dec 22, 2025Abstract:Continual self-supervised learning (CSSL) in medical imaging trains a foundation model sequentially, alleviating the need for collecting multi-modal images for joint training and offering promising improvements in downstream performance while preserving data privacy. However, most existing methods still rely on replaying data from previous stages to prevent catastrophic forgetting, which compromises privacy and limits their applicability in real-world scenarios where data transfer across sites is often restricted. In this work, we propose InvCoSS, an inversion-driven continual self-supervised learning framework for medical multi-modal image pre-training. Specifically, after training on a previous task, InvCoSS inverts the pre-trained self-supervised model to generate synthetic images that approximate the original training distribution. These synthetic images are then combined with data from the new task for joint optimization, which effectively mitigates catastrophic forgetting while strictly adhering to the constraint of no access to previous real data. Furthermore, to improve the fidelity of synthetic images, we introduce a novel InvUNet with a multi-scale fusion architecture to restore both high- and low-frequency components of the inverted images. To enhance diversity and prevent mode collapse, we design a repulsive representation-learning mechanism that encourages a diverse feature space for synthetic images without class guidance. Extensive experiments across nine downstream tasks validate the effectiveness of InvCoSS, achieving performance comparable to or even superior to prior data-replay methods while significantly reducing storage requirements and eliminating data privacy constraints.
DINOv3-Guided Cross Fusion Framework for Semantic-aware CT generation from MRI and CBCT
Nov 15, 2025



Abstract:Generating synthetic CT images from CBCT or MRI has a potential for efficient radiation dose planning and adaptive radiotherapy. However, existing CNN-based models lack global semantic understanding, while Transformers often overfit small medical datasets due to high model capacity and weak inductive bias. To address these limitations, we propose a DINOv3-Guided Cross Fusion (DGCF) framework that integrates a frozen self-supervised DINOv3 Transformer with a trainable CNN encoder-decoder. It hierarchically fuses global representation of Transformer and local features of CNN via a learnable cross fusion module, achieving balanced local appearance and contextual representation. Furthermore, we introduce a Multi-Level DINOv3 Perceptual (MLDP) loss that encourages semantic similarity between synthetic CT and the ground truth in DINOv3's feature space. Experiments on the SynthRAD2023 pelvic dataset demonstrate that DGCF achieved state-of-the-art performance in terms of MS-SSIM, PSNR and segmentation-based metrics on both MRI$\rightarrow$CT and CBCT$\rightarrow$CT translation tasks. To the best of our knowledge, this is the first work to employ DINOv3 representations for medical image translation, highlighting the potential of self-supervised Transformer guidance for semantic-aware CT synthesis. The code is available at https://github.com/HiLab-git/DGCF.
OpenPath: Open-Set Active Learning for Pathology Image Classification via Pre-trained Vision-Language Models
Jun 18, 2025Abstract:Pathology image classification plays a crucial role in accurate medical diagnosis and treatment planning. Training high-performance models for this task typically requires large-scale annotated datasets, which are both expensive and time-consuming to acquire. Active Learning (AL) offers a solution by iteratively selecting the most informative samples for annotation, thereby reducing the labeling effort. However, most AL methods are designed under the assumption of a closed-set scenario, where all the unannotated images belong to target classes. In real-world clinical environments, the unlabeled pool often contains a substantial amount of Out-Of-Distribution (OOD) data, leading to low efficiency of annotation in traditional AL methods. Furthermore, most existing AL methods start with random selection in the first query round, leading to a significant waste of labeling costs in open-set scenarios. To address these challenges, we propose OpenPath, a novel open-set active learning approach for pathological image classification leveraging a pre-trained Vision-Language Model (VLM). In the first query, we propose task-specific prompts that combine target and relevant non-target class prompts to effectively select In-Distribution (ID) and informative samples from the unlabeled pool. In subsequent queries, Diverse Informative ID Sampling (DIS) that includes Prototype-based ID candidate Selection (PIS) and Entropy-Guided Stochastic Sampling (EGSS) is proposed to ensure both purity and informativeness in a query, avoiding the selection of OOD samples. Experiments on two public pathology image datasets show that OpenPath significantly enhances the model's performance due to its high purity of selected samples, and outperforms several state-of-the-art open-set AL methods. The code is available at \href{https://github.com/HiLab-git/OpenPath}{https://github.com/HiLab-git/OpenPath}..
SRPL-SFDA: SAM-Guided Reliable Pseudo-Labels for Source-Free Domain Adaptation in Medical Image Segmentation
Jun 11, 2025Abstract:Domain Adaptation (DA) is crucial for robust deployment of medical image segmentation models when applied to new clinical centers with significant domain shifts. Source-Free Domain Adaptation (SFDA) is appealing as it can deal with privacy concerns and access constraints on source-domain data during adaptation to target-domain data. However, SFDA faces challenges such as insufficient supervision in the target domain with unlabeled images. In this work, we propose a Segment Anything Model (SAM)-guided Reliable Pseudo-Labels method for SFDA (SRPL-SFDA) with three key components: 1) Test-Time Tri-branch Intensity Enhancement (T3IE) that not only improves quality of raw pseudo-labels in the target domain, but also leads to SAM-compatible inputs with three channels to better leverage SAM's zero-shot inference ability for refining the pseudo-labels; 2) A reliable pseudo-label selection module that rejects low-quality pseudo-labels based on Consistency of Multiple SAM Outputs (CMSO) under input perturbations with T3IE; and 3) A reliability-aware training procedure in the unlabeled target domain where reliable pseudo-labels are used for supervision and unreliable parts are regularized by entropy minimization. Experiments conducted on two multi-domain medical image segmentation datasets for fetal brain and the prostate respectively demonstrate that: 1) SRPL-SFDA effectively enhances pseudo-label quality in the unlabeled target domain, and improves SFDA performance by leveraging the reliability-aware training; 2) SRPL-SFDA outperformed state-of-the-art SFDA methods, and its performance is close to that of supervised training in the target domain. The code of this work is available online: https://github.com/HiLab-git/SRPL-SFDA.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025


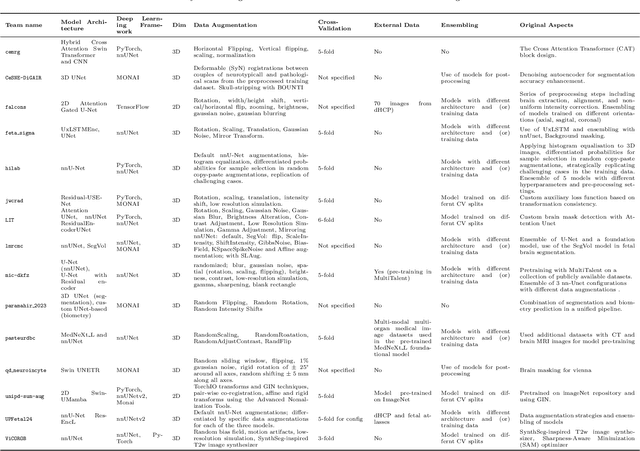
Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
Fairness Analysis of CLIP-Based Foundation Models for X-Ray Image Classification
Jan 31, 2025Abstract:X-ray imaging is pivotal in medical diagnostics, offering non-invasive insights into a range of health conditions. Recently, vision-language models, such as the Contrastive Language-Image Pretraining (CLIP) model, have demonstrated potential in improving diagnostic accuracy by leveraging large-scale image-text datasets. However, since CLIP was not initially designed for medical images, several CLIP-like models trained specifically on medical images have been developed. Despite their enhanced performance, issues of fairness - particularly regarding demographic attributes - remain largely unaddressed. In this study, we perform a comprehensive fairness analysis of CLIP-like models applied to X-ray image classification. We assess their performance and fairness across diverse patient demographics and disease categories using zero-shot inference and various fine-tuning techniques, including Linear Probing, Multilayer Perceptron (MLP), Low-Rank Adaptation (LoRA), and full fine-tuning. Our results indicate that while fine-tuning improves model accuracy, fairness concerns persist, highlighting the need for further fairness interventions in these foundational models.
CLISC: Bridging clip and sam by enhanced cam for unsupervised brain tumor segmentation
Jan 27, 2025



Abstract:Brain tumor segmentation is important for diagnosis of the tumor, and current deep-learning methods rely on a large set of annotated images for training, with high annotation costs. Unsupervised segmentation is promising to avoid human annotations while the performance is often limited. In this study, we present a novel unsupervised segmentation approach that leverages the capabilities of foundation models, and it consists of three main steps: (1) A vision-language model (i.e., CLIP) is employed to obtain image-level pseudo-labels for training a classification network. Class Activation Mapping (CAM) is then employed to extract Regions of Interest (ROIs), where an adaptive masking-based data augmentation is used to enhance ROI identification.(2) The ROIs are used to generate bounding box and point prompts for the Segment Anything Model (SAM) to obtain segmentation pseudo-labels. (3) A 3D segmentation network is trained with the SAM-derived pseudo-labels, where low-quality pseudo-labels are filtered out in a self-learning process based on the similarity between the SAM's output and the network's prediction. Evaluation on the BraTS2020 dataset demonstrates that our approach obtained an average Dice Similarity Score (DSC) of 85.60%, outperforming five state-of-the-art unsupervised segmentation methods by more than 10 percentage points. Besides, our approach outperforms directly using SAM for zero-shot inference, and its performance is close to fully supervised learning.
GLFC: Unified Global-Local Feature and Contrast Learning with Mamba-Enhanced UNet for Synthetic CT Generation from CBCT
Jan 06, 2025



Abstract:Generating synthetic Computed Tomography (CT) images from Cone Beam Computed Tomography (CBCT) is desirable for improving the image quality of CBCT. Existing synthetic CT (sCT) generation methods using Convolutional Neural Networks (CNN) and Transformers often face difficulties in effectively capturing both global and local features and contrasts for high-quality sCT generation. In this work, we propose a Global-Local Feature and Contrast learning (GLFC) framework for sCT generation. First, a Mamba-Enhanced UNet (MEUNet) is introduced by integrating Mamba blocks into the skip connections of a high-resolution UNet for effective global and local feature learning. Second, we propose a Multiple Contrast Loss (MCL) that calculates synthetic loss at different intensity windows to improve quality for both soft tissues and bone regions. Experiments on the SynthRAD2023 dataset demonstrate that GLFC improved the SSIM of sCT from 77.91% to 91.50% compared with the original CBCT, and significantly outperformed several existing methods for sCT generation. The code is available at https://github.com/intelland/GLFC
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge