Yuyin Zhou
OpenVision 3: A Family of Unified Visual Encoder for Both Understanding and Generation
Jan 21, 2026Abstract:This paper presents a family of advanced vision encoder, named OpenVision 3, that learns a single, unified visual representation that can serve both image understanding and image generation. Our core architecture is simple: we feed VAE-compressed image latents to a ViT encoder and train its output to support two complementary roles. First, the encoder output is passed to the ViT-VAE decoder to reconstruct the original image, encouraging the representation to capture generative structure. Second, the same representation is optimized with contrastive learning and image-captioning objectives, strengthening semantic features. By jointly optimizing reconstruction- and semantics-driven signals in a shared latent space, the encoder learns representations that synergize and generalize well across both regimes. We validate this unified design through extensive downstream evaluations with the encoder frozen. For multimodal understanding, we plug the encoder into the LLaVA-1.5 framework: it performs comparably with a standard CLIP vision encoder (e.g., 62.4 vs 62.2 on SeedBench, and 83.7 vs 82.9 on POPE). For generation, we test it under the RAE framework: ours substantially surpasses the standard CLIP-based encoder (e.g., gFID: 1.89 vs 2.54 on ImageNet). We hope this work can spur future research on unified modeling.
Controllable Layered Image Generation for Real-World Editing
Jan 21, 2026Abstract:Recent image generation models have shown impressive progress, yet they often struggle to yield controllable and consistent results when users attempt to edit specific elements within an existing image. Layered representations enable flexible, user-driven content creation, but existing approaches often fail to produce layers with coherent compositing relationships, and their object layers typically lack realistic visual effects such as shadows and reflections. To overcome these limitations, we propose LASAGNA, a novel, unified framework that generates an image jointly with its composing layers--a photorealistic background and a high-quality transparent foreground with compelling visual effects. Unlike prior work, LASAGNA efficiently learns correct image composition from a wide range of conditioning inputs--text prompts, foreground, background, and location masks--offering greater controllability for real-world applications. To enable this, we introduce LASAGNA-48K, a new dataset composed of clean backgrounds and RGBA foregrounds with physically grounded visual effects. We also propose LASAGNABENCH, the first benchmark for layer editing. We demonstrate that LASAGNA excels in generating highly consistent and coherent results across multiple image layers simultaneously, enabling diverse post-editing applications that accurately preserve identity and visual effects. LASAGNA-48K and LASAGNABENCH will be publicly released to foster open research in the community. The project page is https://rayjryang.github.io/LASAGNA-Page/.
All You Need Are Random Visual Tokens? Demystifying Token Pruning in VLLMs
Dec 08, 2025Abstract:Vision Large Language Models (VLLMs) incur high computational costs due to their reliance on hundreds of visual tokens to represent images. While token pruning offers a promising solution for accelerating inference, this paper, however, identifies a key observation: in deeper layers (e.g., beyond the 20th), existing training-free pruning methods perform no better than random pruning. We hypothesize that this degradation is caused by "vanishing token information", where visual tokens progressively lose their salience with increasing network depth. To validate this hypothesis, we quantify a token's information content by measuring the change in the model output probabilities upon its removal. Using this proposed metric, our analysis of the information of visual tokens across layers reveals three key findings: (1) As layers deepen, the information of visual tokens gradually becomes uniform and eventually vanishes at an intermediate layer, which we term as "information horizon", beyond which the visual tokens become redundant; (2) The position of this horizon is not static; it extends deeper for visually intensive tasks, such as Optical Character Recognition (OCR), compared to more general tasks like Visual Question Answering (VQA); (3) This horizon is also strongly correlated with model capacity, as stronger VLLMs (e.g., Qwen2.5-VL) employ deeper visual tokens than weaker models (e.g., LLaVA-1.5). Based on our findings, we show that simple random pruning in deep layers efficiently balances performance and efficiency. Moreover, integrating random pruning consistently enhances existing methods. Using DivPrune with random pruning achieves state-of-the-art results, maintaining 96.9% of Qwen-2.5-VL-7B performance while pruning 50% of visual tokens. The code will be publicly available at https://github.com/YahongWang1/Information-Horizon.
MedVLSynther: Synthesizing High-Quality Visual Question Answering from Medical Documents with Generator-Verifier LMMs
Oct 29, 2025Abstract:Large Multimodal Models (LMMs) are increasingly capable of answering medical questions that require joint reasoning over images and text, yet training general medical VQA systems is impeded by the lack of large, openly usable, high-quality corpora. We present MedVLSynther, a rubric-guided generator-verifier framework that synthesizes high-quality multiple-choice VQA items directly from open biomedical literature by conditioning on figures, captions, and in-text references. The generator produces self-contained stems and parallel, mutually exclusive options under a machine-checkable JSON schema; a multi-stage verifier enforces essential gates (self-containment, single correct answer, clinical validity, image-text consistency), awards fine-grained positive points, and penalizes common failure modes before acceptance. Applying this pipeline to PubMed Central yields MedSynVQA: 13,087 audited questions over 14,803 images spanning 13 imaging modalities and 28 anatomical regions. Training open-weight LMMs with reinforcement learning using verifiable rewards improves accuracy across six medical VQA benchmarks, achieving averages of 55.85 (3B) and 58.15 (7B), with up to 77.57 on VQA-RAD and 67.76 on PathVQA, outperforming strong medical LMMs. A Ablations verify that both generation and verification are necessary and that more verified data consistently helps, and a targeted contamination analysis detects no leakage from evaluation suites. By operating entirely on open literature and open-weight models, MedVLSynther offers an auditable, reproducible, and privacy-preserving path to scalable medical VQA training data.
GauSSmart: Enhanced 3D Reconstruction through 2D Foundation Models and Geometric Filtering
Oct 16, 2025Abstract:Scene reconstruction has emerged as a central challenge in computer vision, with approaches such as Neural Radiance Fields (NeRF) and Gaussian Splatting achieving remarkable progress. While Gaussian Splatting demonstrates strong performance on large-scale datasets, it often struggles to capture fine details or maintain realism in regions with sparse coverage, largely due to the inherent limitations of sparse 3D training data. In this work, we propose GauSSmart, a hybrid method that effectively bridges 2D foundational models and 3D Gaussian Splatting reconstruction. Our approach integrates established 2D computer vision techniques, including convex filtering and semantic feature supervision from foundational models such as DINO, to enhance Gaussian-based scene reconstruction. By leveraging 2D segmentation priors and high-dimensional feature embeddings, our method guides the densification and refinement of Gaussian splats, improving coverage in underrepresented areas and preserving intricate structural details. We validate our approach across three datasets, where GauSSmart consistently outperforms existing Gaussian Splatting in the majority of evaluated scenes. Our results demonstrate the significant potential of hybrid 2D-3D approaches, highlighting how the thoughtful combination of 2D foundational models with 3D reconstruction pipelines can overcome the limitations inherent in either approach alone.
GPT-IMAGE-EDIT-1.5M: A Million-Scale, GPT-Generated Image Dataset
Jul 28, 2025
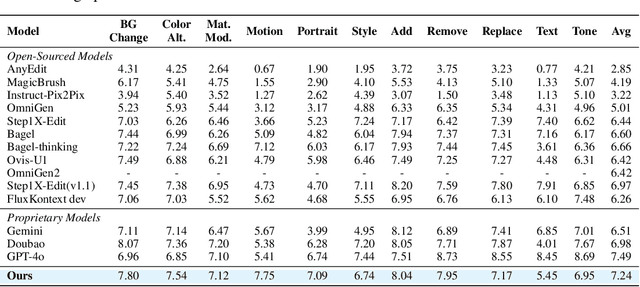
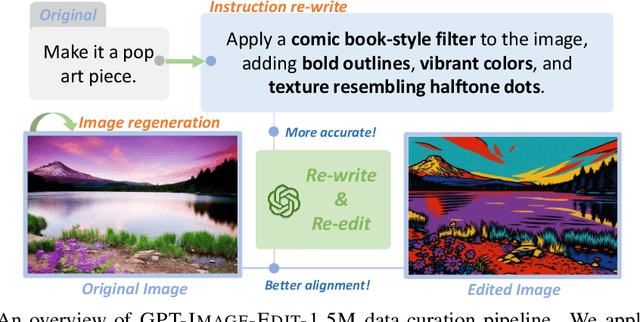
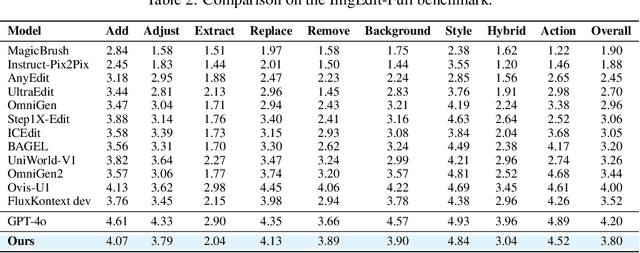
Abstract:Recent advancements in large multimodal models like GPT-4o have set a new standard for high-fidelity, instruction-guided image editing. However, the proprietary nature of these models and their training data creates a significant barrier for open-source research. To bridge this gap, we introduce GPT-IMAGE-EDIT-1.5M, a publicly available, large-scale image-editing corpus containing more than 1.5 million high-quality triplets (instruction, source image, edited image). We systematically construct this dataset by leveraging the versatile capabilities of GPT-4o to unify and refine three popular image-editing datasets: OmniEdit, HQ-Edit, and UltraEdit. Specifically, our methodology involves 1) regenerating output images to enhance visual quality and instruction alignment, and 2) selectively rewriting prompts to improve semantic clarity. To validate the efficacy of our dataset, we fine-tune advanced open-source models on GPT-IMAGE-EDIT-1.5M. The empirical results are exciting, e.g., the fine-tuned FluxKontext achieves highly competitive performance across a comprehensive suite of benchmarks, including 7.24 on GEdit-EN, 3.80 on ImgEdit-Full, and 8.78 on Complex-Edit, showing stronger instruction following and higher perceptual quality while maintaining identity. These scores markedly exceed all previously published open-source methods and substantially narrow the gap to leading proprietary models. We hope the full release of GPT-IMAGE-EDIT-1.5M can help to catalyze further open research in instruction-guided image editing.
A Survey on Latent Reasoning
Jul 08, 2025



Abstract:Large Language Models (LLMs) have demonstrated impressive reasoning capabilities, especially when guided by explicit chain-of-thought (CoT) reasoning that verbalizes intermediate steps. While CoT improves both interpretability and accuracy, its dependence on natural language reasoning limits the model's expressive bandwidth. Latent reasoning tackles this bottleneck by performing multi-step inference entirely in the model's continuous hidden state, eliminating token-level supervision. To advance latent reasoning research, this survey provides a comprehensive overview of the emerging field of latent reasoning. We begin by examining the foundational role of neural network layers as the computational substrate for reasoning, highlighting how hierarchical representations support complex transformations. Next, we explore diverse latent reasoning methodologies, including activation-based recurrence, hidden state propagation, and fine-tuning strategies that compress or internalize explicit reasoning traces. Finally, we discuss advanced paradigms such as infinite-depth latent reasoning via masked diffusion models, which enable globally consistent and reversible reasoning processes. By unifying these perspectives, we aim to clarify the conceptual landscape of latent reasoning and chart future directions for research at the frontier of LLM cognition. An associated GitHub repository collecting the latest papers and repos is available at: https://github.com/multimodal-art-projection/LatentCoT-Horizon/.
FedVLMBench: Benchmarking Federated Fine-Tuning of Vision-Language Models
Jun 11, 2025Abstract:Vision-Language Models (VLMs) have demonstrated remarkable capabilities in cross-modal understanding and generation by integrating visual and textual information. While instruction tuning and parameter-efficient fine-tuning methods have substantially improved the generalization of VLMs, most existing approaches rely on centralized training, posing challenges for deployment in domains with strict privacy requirements like healthcare. Recent efforts have introduced Federated Learning (FL) into VLM fine-tuning to address these privacy concerns, yet comprehensive benchmarks for evaluating federated fine-tuning strategies, model architectures, and task generalization remain lacking. In this work, we present \textbf{FedVLMBench}, the first systematic benchmark for federated fine-tuning of VLMs. FedVLMBench integrates two mainstream VLM architectures (encoder-based and encoder-free), four fine-tuning strategies, five FL algorithms, six multimodal datasets spanning four cross-domain single-task scenarios and two cross-domain multitask settings, covering four distinct downstream task categories. Through extensive experiments, we uncover key insights into the interplay between VLM architectures, fine-tuning strategies, data heterogeneity, and multi-task federated optimization. Notably, we find that a 2-layer multilayer perceptron (MLP) connector with concurrent connector and LLM tuning emerges as the optimal configuration for encoder-based VLMs in FL. Furthermore, current FL methods exhibit significantly higher sensitivity to data heterogeneity in vision-centric tasks than text-centric ones, across both encoder-free and encoder-based VLM architectures. Our benchmark provides essential tools, datasets, and empirical guidance for the research community, offering a standardized platform to advance privacy-preserving, federated training of multimodal foundation models.
Harnessing EHRs for Diffusion-based Anomaly Detection on Chest X-rays
May 22, 2025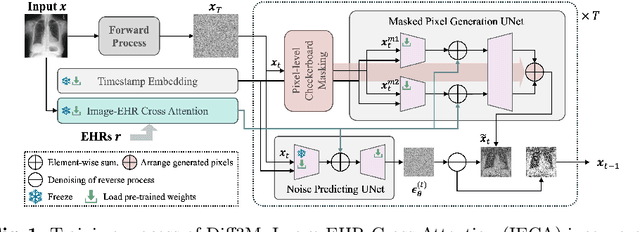
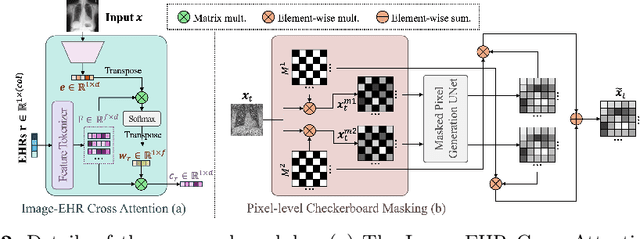
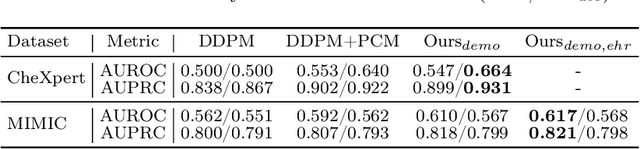
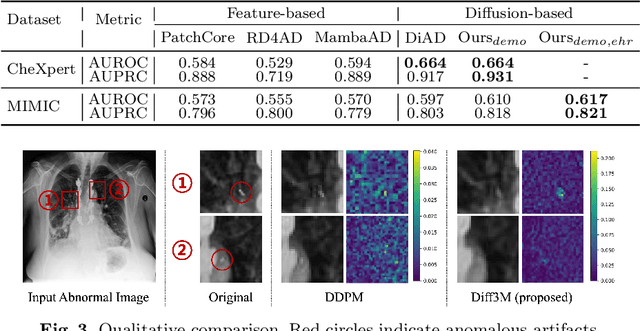
Abstract:Unsupervised anomaly detection (UAD) in medical imaging is crucial for identifying pathological abnormalities without requiring extensive labeled data. However, existing diffusion-based UAD models rely solely on imaging features, limiting their ability to distinguish between normal anatomical variations and pathological anomalies. To address this, we propose Diff3M, a multi-modal diffusion-based framework that integrates chest X-rays and structured Electronic Health Records (EHRs) for enhanced anomaly detection. Specifically, we introduce a novel image-EHR cross-attention module to incorporate structured clinical context into the image generation process, improving the model's ability to differentiate normal from abnormal features. Additionally, we develop a static masking strategy to enhance the reconstruction of normal-like images from anomalies. Extensive evaluations on CheXpert and MIMIC-CXR/IV demonstrate that Diff3M achieves state-of-the-art performance, outperforming existing UAD methods in medical imaging. Our code is available at this http URL https://github.com/nth221/Diff3M
ATR-Bench: A Federated Learning Benchmark for Adaptation, Trust, and Reasoning
May 22, 2025Abstract:Federated Learning (FL) has emerged as a promising paradigm for collaborative model training while preserving data privacy across decentralized participants. As FL adoption grows, numerous techniques have been proposed to tackle its practical challenges. However, the lack of standardized evaluation across key dimensions hampers systematic progress and fair comparison of FL methods. In this work, we introduce ATR-Bench, a unified framework for analyzing federated learning through three foundational dimensions: Adaptation, Trust, and Reasoning. We provide an in-depth examination of the conceptual foundations, task formulations, and open research challenges associated with each theme. We have extensively benchmarked representative methods and datasets for adaptation to heterogeneous clients and trustworthiness in adversarial or unreliable environments. Due to the lack of reliable metrics and models for reasoning in FL, we only provide literature-driven insights for this dimension. ATR-Bench lays the groundwork for a systematic and holistic evaluation of federated learning with real-world relevance. We will make our complete codebase publicly accessible and a curated repository that continuously tracks new developments and research in the FL literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge