Yinan Chen
UltraVideo: High-Quality UHD Video Dataset with Comprehensive Captions
Jun 16, 2025Abstract:The quality of the video dataset (image quality, resolution, and fine-grained caption) greatly influences the performance of the video generation model. The growing demand for video applications sets higher requirements for high-quality video generation models. For example, the generation of movie-level Ultra-High Definition (UHD) videos and the creation of 4K short video content. However, the existing public datasets cannot support related research and applications. In this paper, we first propose a high-quality open-sourced UHD-4K (22.4\% of which are 8K) text-to-video dataset named UltraVideo, which contains a wide range of topics (more than 100 kinds), and each video has 9 structured captions with one summarized caption (average of 824 words). Specifically, we carefully design a highly automated curation process with four stages to obtain the final high-quality dataset: \textit{i)} collection of diverse and high-quality video clips. \textit{ii)} statistical data filtering. \textit{iii)} model-based data purification. \textit{iv)} generation of comprehensive, structured captions. In addition, we expand Wan to UltraWan-1K/-4K, which can natively generate high-quality 1K/4K videos with more consistent text controllability, demonstrating the effectiveness of our data curation.We believe that this work can make a significant contribution to future research on UHD video generation. UltraVideo dataset and UltraWan models are available at https://xzc-zju.github.io/projects/UltraVideo.
Multi-Prototype Embedding Refinement for Semi-Supervised Medical Image Segmentation
Mar 18, 2025



Abstract:Medical image segmentation aims to identify anatomical structures at the voxel-level. Segmentation accuracy relies on distinguishing voxel differences. Compared to advancements achieved in studies of the inter-class variance, the intra-class variance receives less attention. Moreover, traditional linear classifiers, limited by a single learnable weight per class, struggle to capture this finer distinction. To address the above challenges, we propose a Multi-Prototype-based Embedding Refinement method for semi-supervised medical image segmentation. Specifically, we design a multi-prototype-based classification strategy, rethinking the segmentation from the perspective of structural relationships between voxel embeddings. The intra-class variations are explored by clustering voxels along the distribution of multiple prototypes in each class. Next, we introduce a consistency constraint to alleviate the limitation of linear classifiers. This constraint integrates different classification granularities from a linear classifier and the proposed prototype-based classifier. In the thorough evaluation on two popular benchmarks, our method achieves superior performance compared with state-of-the-art methods. Code is available at https://github.com/Briley-byl123/MPER.
Image Inversion: A Survey from GANs to Diffusion and Beyond
Feb 17, 2025Abstract:Image inversion is a fundamental task in generative models, aiming to map images back to their latent representations to enable downstream applications such as editing, restoration, and style transfer. This paper provides a comprehensive review of the latest advancements in image inversion techniques, focusing on two main paradigms: Generative Adversarial Network (GAN) inversion and diffusion model inversion. We categorize these techniques based on their optimization methods. For GAN inversion, we systematically classify existing methods into encoder-based approaches, latent optimization approaches, and hybrid approaches, analyzing their theoretical foundations, technical innovations, and practical trade-offs. For diffusion model inversion, we explore training-free strategies, fine-tuning methods, and the design of additional trainable modules, highlighting their unique advantages and limitations. Additionally, we discuss several popular downstream applications and emerging applications beyond image tasks, identifying current challenges and future research directions. By synthesizing the latest developments, this paper aims to provide researchers and practitioners with a valuable reference resource, promoting further advancements in the field of image inversion. We keep track of the latest works at https://github.com/RyanChenYN/ImageInversion
DiffSeg: A Segmentation Model for Skin Lesions Based on Diffusion Difference
Apr 25, 2024



Abstract:Weakly supervised medical image segmentation (MIS) using generative models is crucial for clinical diagnosis. However, the accuracy of the segmentation results is often limited by insufficient supervision and the complex nature of medical imaging. Existing models also only provide a single outcome, which does not allow for the measurement of uncertainty. In this paper, we introduce DiffSeg, a segmentation model for skin lesions based on diffusion difference which exploits diffusion model principles to ex-tract noise-based features from images with diverse semantic information. By discerning difference between these noise features, the model identifies diseased areas. Moreover, its multi-output capability mimics doctors' annotation behavior, facilitating the visualization of segmentation result consistency and ambiguity. Additionally, it quantifies output uncertainty using Generalized Energy Distance (GED), aiding interpretability and decision-making for physicians. Finally, the model integrates outputs through the Dense Conditional Random Field (DenseCRF) algorithm to refine the segmentation boundaries by considering inter-pixel correlations, which improves the accuracy and optimizes the segmentation results. We demonstrate the effectiveness of DiffSeg on the ISIC 2018 Challenge dataset, outperforming state-of-the-art U-Net-based methods.
A Sentiment Analysis of Medical Text Based on Deep Learning
Apr 16, 2024



Abstract:The field of natural language processing (NLP) has made significant progress with the rapid development of deep learning technologies. One of the research directions in text sentiment analysis is sentiment analysis of medical texts, which holds great potential for application in clinical diagnosis. However, the medical field currently lacks sufficient text datasets, and the effectiveness of sentiment analysis is greatly impacted by different model design approaches, which presents challenges. Therefore, this paper focuses on the medical domain, using bidirectional encoder representations from transformers (BERT) as the basic pre-trained model and experimenting with modules such as convolutional neural network (CNN), fully connected network (FCN), and graph convolutional networks (GCN) at the output layer. Experiments and analyses were conducted on the METS-CoV dataset to explore the training performance after integrating different deep learning networks. The results indicate that CNN models outperform other networks when trained on smaller medical text datasets in combination with pre-trained models like BERT. This study highlights the significance of model selection in achieving effective sentiment analysis in the medical domain and provides a reference for future research to develop more efficient model architectures.
UPL-SFDA: Uncertainty-aware Pseudo Label Guided Source-Free Domain Adaptation for Medical Image Segmentation
Sep 19, 2023



Abstract:Domain Adaptation (DA) is important for deep learning-based medical image segmentation models to deal with testing images from a new target domain. As the source-domain data are usually unavailable when a trained model is deployed at a new center, Source-Free Domain Adaptation (SFDA) is appealing for data and annotation-efficient adaptation to the target domain. However, existing SFDA methods have a limited performance due to lack of sufficient supervision with source-domain images unavailable and target-domain images unlabeled. We propose a novel Uncertainty-aware Pseudo Label guided (UPL) SFDA method for medical image segmentation. Specifically, we propose Target Domain Growing (TDG) to enhance the diversity of predictions in the target domain by duplicating the pre-trained model's prediction head multiple times with perturbations. The different predictions in these duplicated heads are used to obtain pseudo labels for unlabeled target-domain images and their uncertainty to identify reliable pseudo labels. We also propose a Twice Forward pass Supervision (TFS) strategy that uses reliable pseudo labels obtained in one forward pass to supervise predictions in the next forward pass. The adaptation is further regularized by a mean prediction-based entropy minimization term that encourages confident and consistent results in different prediction heads. UPL-SFDA was validated with a multi-site heart MRI segmentation dataset, a cross-modality fetal brain segmentation dataset, and a 3D fetal tissue segmentation dataset. It improved the average Dice by 5.54, 5.01 and 6.89 percentage points for the three tasks compared with the baseline, respectively, and outperformed several state-of-the-art SFDA methods.
Automatic lobe segmentation using attentive cross entropy and end-to-end fissure generation
Jul 24, 2023



Abstract:The automatic lung lobe segmentation algorithm is of great significance for the diagnosis and treatment of lung diseases, however, which has great challenges due to the incompleteness of pulmonary fissures in lung CT images and the large variability of pathological features. Therefore, we propose a new automatic lung lobe segmentation framework, in which we urge the model to pay attention to the area around the pulmonary fissure during the training process, which is realized by a task-specific loss function. In addition, we introduce an end-to-end pulmonary fissure generation method in the auxiliary pulmonary fissure segmentation task, without any additional network branch. Finally, we propose a registration-based loss function to alleviate the convergence difficulty of the Dice loss supervised pulmonary fissure segmentation task. We achieve 97.83% and 94.75% dice scores on our private dataset STLB and public LUNA16 dataset respectively.
Transforming Graphs for Enhanced Attribute-Based Clustering: An Innovative Graph Transformer Method
Jun 21, 2023



Abstract:Graph Representation Learning (GRL) is an influential methodology, enabling a more profound understanding of graph-structured data and aiding graph clustering, a critical task across various domains. The recent incursion of attention mechanisms, originally an artifact of Natural Language Processing (NLP), into the realm of graph learning has spearheaded a notable shift in research trends. Consequently, Graph Attention Networks (GATs) and Graph Attention Auto-Encoders have emerged as preferred tools for graph clustering tasks. Yet, these methods primarily employ a local attention mechanism, thereby curbing their capacity to apprehend the intricate global dependencies between nodes within graphs. Addressing these impediments, this study introduces an innovative method known as the Graph Transformer Auto-Encoder for Graph Clustering (GTAGC). By melding the Graph Auto-Encoder with the Graph Transformer, GTAGC is adept at capturing global dependencies between nodes. This integration amplifies the graph representation and surmounts the constraints posed by the local attention mechanism. The architecture of GTAGC encompasses graph embedding, integration of the Graph Transformer within the autoencoder structure, and a clustering component. It strategically alternates between graph embedding and clustering, thereby tailoring the Graph Transformer for clustering tasks, whilst preserving the graph's global structural information. Through extensive experimentation on diverse benchmark datasets, GTAGC has exhibited superior performance against existing state-of-the-art graph clustering methodologies. This pioneering approach represents a novel contribution to the field of graph clustering, paving the way for promising avenues in future research.
CDDSA: Contrastive Domain Disentanglement and Style Augmentation for Generalizable Medical Image Segmentation
Nov 22, 2022



Abstract:Generalization to previously unseen images with potential domain shifts and different styles is essential for clinically applicable medical image segmentation, and the ability to disentangle domain-specific and domain-invariant features is key for achieving Domain Generalization (DG). However, existing DG methods can hardly achieve effective disentanglement to get high generalizability. To deal with this problem, we propose an efficient Contrastive Domain Disentanglement and Style Augmentation (CDDSA) framework for generalizable medical image segmentation. First, a disentangle network is proposed to decompose an image into a domain-invariant anatomical representation and a domain-specific style code, where the former is sent to a segmentation model that is not affected by the domain shift, and the disentangle network is regularized by a decoder that combines the anatomical and style codes to reconstruct the input image. Second, to achieve better disentanglement, a contrastive loss is proposed to encourage the style codes from the same domain and different domains to be compact and divergent, respectively. Thirdly, to further improve generalizability, we propose a style augmentation method based on the disentanglement representation to synthesize images in various unseen styles with shared anatomical structures. Our method was validated on a public multi-site fundus image dataset for optic cup and disc segmentation and an in-house multi-site Nasopharyngeal Carcinoma Magnetic Resonance Image (NPC-MRI) dataset for nasopharynx Gross Tumor Volume (GTVnx) segmentation. Experimental results showed that the proposed CDDSA achieved remarkable generalizability across different domains, and it outperformed several state-of-the-art methods in domain-generalizable segmentation.
SCPM-Net: An Anchor-free 3D Lung Nodule Detection Network using Sphere Representation and Center Points Matching
Apr 12, 2021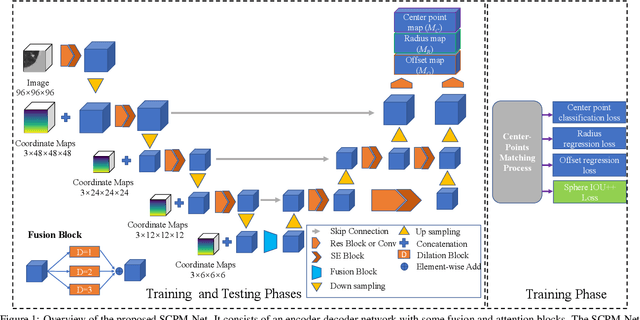
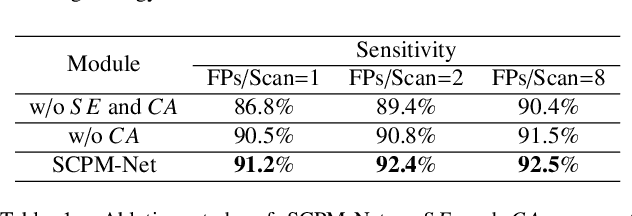
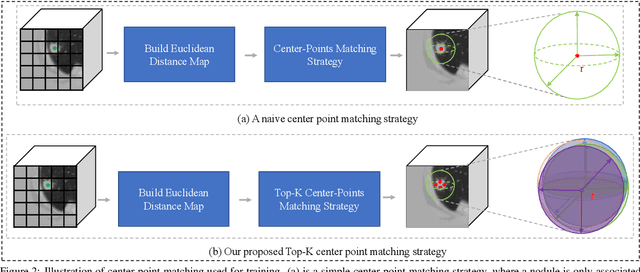
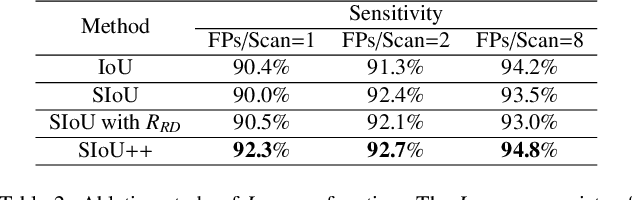
Abstract:Automatic and accurate lung nodule detection from 3D Computed Tomography scans plays a vital role in efficient lung cancer screening. Despite the state-of-the-art performance obtained by recent anchor-based detectors using Convolutional Neural Networks, they require predetermined anchor parameters such as the size, number, and aspect ratio of anchors, and have limited robustness when dealing with lung nodules with a massive variety of sizes. We propose a 3D sphere representation-based center-points matching detection network (SCPM-Net) that is anchor-free and automatically predicts the position, radius, and offset of nodules without the manual design of nodule/anchor parameters. The SCPM-Net consists of two novel pillars: sphere representation and center points matching. To mimic the nodule annotation in clinical practice, we replace the conventional bounding box with the newly proposed bounding sphere. A compatible sphere-based intersection over-union loss function is introduced to train the lung nodule detection network stably and efficiently.We empower the network anchor-free by designing a positive center-points selection and matching (CPM) process, which naturally discards pre-determined anchor boxes. An online hard example mining and re-focal loss subsequently enable the CPM process more robust, resulting in more accurate point assignment and the mitigation of class imbalance. In addition, to better capture spatial information and 3D context for the detection, we propose to fuse multi-level spatial coordinate maps with the feature extractor and combine them with 3D squeeze-and-excitation attention modules. Experimental results on the LUNA16 dataset showed that our proposed SCPM-Net framework achieves superior performance compared with existing used anchor-based and anchor-free methods for lung nodule detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge