Ehsan Adeli
Department of Computer Science, Stanford University, Stanford, CA, USA, Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, CA, USA
Anatomically Guided Latent Diffusion for Brain MRI Progression Modeling
Jan 21, 2026Abstract:Accurately modeling longitudinal brain MRI progression is crucial for understanding neurodegenerative diseases and predicting individualized structural changes. Existing state-of-the-art approaches, such as Brain Latent Progression (BrLP), often use multi-stage training pipelines with auxiliary conditioning modules but suffer from architectural complexity, suboptimal use of conditional clinical covariates, and limited guarantees of anatomical consistency. We propose Anatomically Guided Latent Diffusion Model (AG-LDM), a segmentation-guided framework that enforces anatomically consistent progression while substantially simplifying the training pipeline. AG-LDM conditions latent diffusion by directly fusing baseline anatomy, noisy follow-up states, and clinical covariates at the input level, a strategy that avoids auxiliary control networks by learning a unified, end-to-end model that represents both anatomy and progression. A lightweight 3D tissue segmentation model (WarpSeg) provides explicit anatomical supervision during both autoencoder fine-tuning and diffusion model training, ensuring consistent brain tissue boundaries and morphometric fidelity. Experiments on 31,713 ADNI longitudinal pairs and zero-shot evaluation on OASIS-3 demonstrate that AG-LDM matches or surpasses more complex diffusion models, achieving state-of-the-art image quality and 15-20\% reduction in volumetric errors in generated images. AG-LDM also exhibits markedly stronger utilization of temporal and clinical covariates (up to 31.5x higher sensitivity than BrLP) and generates biologically plausible counterfactual trajectories, accurately capturing hallmarks of Alzheimer's progression such as limbic atrophy and ventricular expansion. These results highlight AG-LDM as an efficient, anatomically grounded framework for reliable brain MRI progression modeling.
VideoWeave: A Data-Centric Approach for Efficient Video Understanding
Jan 09, 2026Abstract:Training video-language models is often prohibitively expensive due to the high cost of processing long frame sequences and the limited availability of annotated long videos. We present VideoWeave, a simple yet effective approach to improve data efficiency by constructing synthetic long-context training samples that splice together short, captioned videos from existing datasets. Rather than modifying model architectures or optimization objectives, VideoWeave reorganizes available video-text pairs to expand temporal diversity within fixed compute. We systematically study how different data composition strategies like random versus visually clustered splicing and caption enrichment affect downstream performance on downstream video question answering. Under identical compute constraints, models trained with VideoWeave achieve higher accuracy than conventional video finetuning. Our results highlight that reorganizing training data, rather than altering architectures, may offer a simple and scalable path for training video-language models. We link our code for all experiments here.
A Tool Bottleneck Framework for Clinically-Informed and Interpretable Medical Image Understanding
Dec 24, 2025Abstract:Recent tool-use frameworks powered by vision-language models (VLMs) improve image understanding by grounding model predictions with specialized tools. Broadly, these frameworks leverage VLMs and a pre-specified toolbox to decompose the prediction task into multiple tool calls (often deep learning models) which are composed to make a prediction. The dominant approach to composing tools is using text, via function calls embedded in VLM-generated code or natural language. However, these methods often perform poorly on medical image understanding, where salient information is encoded as spatially-localized features that are difficult to compose or fuse via text alone. To address this, we propose a tool-use framework for medical image understanding called the Tool Bottleneck Framework (TBF), which composes VLM-selected tools using a learned Tool Bottleneck Model (TBM). For a given image and task, TBF leverages an off-the-shelf medical VLM to select tools from a toolbox that each extract clinically-relevant features. Instead of text-based composition, these tools are composed by the TBM, which computes and fuses the tool outputs using a neural network before outputting the final prediction. We propose a simple and effective strategy for TBMs to make predictions with any arbitrary VLM tool selection. Overall, our framework not only improves tool-use in medical imaging contexts, but also yields more interpretable, clinically-grounded predictors. We evaluate TBF on tasks in histopathology and dermatology and find that these advantages enable our framework to perform on par with or better than deep learning-based classifiers, VLMs, and state-of-the-art tool-use frameworks, with particular gains in data-limited regimes. Our code is available at https://github.com/christinaliu2020/tool-bottleneck-framework.
QuantiPhy: A Quantitative Benchmark Evaluating Physical Reasoning Abilities of Vision-Language Models
Dec 22, 2025Abstract:Understanding the physical world is essential for generalist AI agents. However, it remains unclear whether state-of-the-art vision perception models (e.g., large VLMs) can reason physical properties quantitatively. Existing evaluations are predominantly VQA-based and qualitative, offering limited insight into whether these models can infer the kinematic quantities of moving objects from video observations. To address this, we present QuantiPhy, the first benchmark designed to quantitatively measure a VLM's physical reasoning ability. Comprising more than 3.3K video-text instances with numerical ground truth, QuantiPhy evaluates a VLM's performance on estimating an object's size, velocity, and acceleration at a given timestamp, using one of these properties as an input prior. The benchmark standardizes prompts and scoring to assess numerical accuracy, enabling fair comparisons across models. Our experiments on state-of-the-art VLMs reveal a consistent gap between their qualitative plausibility and actual numerical correctness. We further provide an in-depth analysis of key factors like background noise, counterfactual priors, and strategic prompting and find that state-of-the-art VLMs lean heavily on pre-trained world knowledge rather than faithfully using the provided visual and textual inputs as references when reasoning kinematic properties quantitatively. QuantiPhy offers the first rigorous, scalable testbed to move VLMs beyond mere verbal plausibility toward a numerically grounded physical understanding.
Repurposing 2D Diffusion Models for 3D Shape Completion
Dec 16, 2025Abstract:We present a framework that adapts 2D diffusion models for 3D shape completion from incomplete point clouds. While text-to-image diffusion models have achieved remarkable success with abundant 2D data, 3D diffusion models lag due to the scarcity of high-quality 3D datasets and a persistent modality gap between 3D inputs and 2D latent spaces. To overcome these limitations, we introduce the Shape Atlas, a compact 2D representation of 3D geometry that (1) enables full utilization of the generative power of pretrained 2D diffusion models, and (2) aligns the modalities between the conditional input and output spaces, allowing more effective conditioning. This unified 2D formulation facilitates learning from limited 3D data and produces high-quality, detail-preserving shape completions. We validate the effectiveness of our results on the PCN and ShapeNet-55 datasets. Additionally, we show the downstream application of creating artist-created meshes from our completed point clouds, further demonstrating the practicality of our method.
ViBES: A Conversational Agent with Behaviorally-Intelligent 3D Virtual Body
Dec 16, 2025



Abstract:Human communication is inherently multimodal and social: words, prosody, and body language jointly carry intent. Yet most prior systems model human behavior as a translation task co-speech gesture or text-to-motion that maps a fixed utterance to motion clips-without requiring agentic decision-making about when to move, what to do, or how to adapt across multi-turn dialogue. This leads to brittle timing, weak social grounding, and fragmented stacks where speech, text, and motion are trained or inferred in isolation. We introduce ViBES (Voice in Behavioral Expression and Synchrony), a conversational 3D agent that jointly plans language and movement and executes dialogue-conditioned body actions. Concretely, ViBES is a speech-language-behavior (SLB) model with a mixture-of-modality-experts (MoME) backbone: modality-partitioned transformer experts for speech, facial expression, and body motion. The model processes interleaved multimodal token streams with hard routing by modality (parameters are split per expert), while sharing information through cross-expert attention. By leveraging strong pretrained speech-language models, the agent supports mixed-initiative interaction: users can speak, type, or issue body-action directives mid-conversation, and the system exposes controllable behavior hooks for streaming responses. We further benchmark on multi-turn conversation with automatic metrics of dialogue-motion alignment and behavior quality, and observe consistent gains over strong co-speech and text-to-motion baselines. ViBES goes beyond "speech-conditioned motion generation" toward agentic virtual bodies where language, prosody, and movement are jointly generated, enabling controllable, socially competent 3D interaction. Code and data will be made available at: ai.stanford.edu/~juze/ViBES/
Spherical Leech Quantization for Visual Tokenization and Generation
Dec 16, 2025



Abstract:Non-parametric quantization has received much attention due to its efficiency on parameters and scalability to a large codebook. In this paper, we present a unified formulation of different non-parametric quantization methods through the lens of lattice coding. The geometry of lattice codes explains the necessity of auxiliary loss terms when training auto-encoders with certain existing lookup-free quantization variants such as BSQ. As a step forward, we explore a few possible candidates, including random lattices, generalized Fibonacci lattices, and densest sphere packing lattices. Among all, we find the Leech lattice-based quantization method, which is dubbed as Spherical Leech Quantization ($Λ_{24}$-SQ), leads to both a simplified training recipe and an improved reconstruction-compression tradeoff thanks to its high symmetry and even distribution on the hypersphere. In image tokenization and compression tasks, this quantization approach achieves better reconstruction quality across all metrics than BSQ, the best prior art, while consuming slightly fewer bits. The improvement also extends to state-of-the-art auto-regressive image generation frameworks.
Ask WhAI:Probing Belief Formation in Role-Primed LLM Agents
Nov 06, 2025Abstract:We present Ask WhAI, a systems-level framework for inspecting and perturbing belief states in multi-agent interactions. The framework records and replays agent interactions, supports out-of-band queries into each agent's beliefs and rationale, and enables counterfactual evidence injection to test how belief structures respond to new information. We apply the framework to a medical case simulator notable for its multi-agent shared memory (a time-stamped electronic medical record, or EMR) and an oracle agent (the LabAgent) that holds ground truth lab results revealed only when explicitly queried. We stress-test the system on a multi-specialty diagnostic journey for a child with an abrupt-onset neuropsychiatric presentation. Large language model agents, each primed with strong role-specific priors ("act like a neurologist", "act like an infectious disease specialist"), write to a shared medical record and interact with a moderator across sequential or parallel encounters. Breakpoints at key diagnostic moments enable pre- and post-event belief queries, allowing us to distinguish entrenched priors from reasoning or evidence-integration effects. The simulation reveals that agent beliefs often mirror real-world disciplinary stances, including overreliance on canonical studies and resistance to counterevidence, and that these beliefs can be traced and interrogated in ways not possible with human experts. By making such dynamics visible and testable, Ask WhAI offers a reproducible way to study belief formation and epistemic silos in multi-agent scientific reasoning.
Artist-Created Mesh Generation from Raw Observation
Sep 15, 2025Abstract:We present an end-to-end framework for generating artist-style meshes from noisy or incomplete point clouds, such as those captured by real-world sensors like LiDAR or mobile RGB-D cameras. Artist-created meshes are crucial for commercial graphics pipelines due to their compatibility with animation and texturing tools and their efficiency in rendering. However, existing approaches often assume clean, complete inputs or rely on complex multi-stage pipelines, limiting their applicability in real-world scenarios. To address this, we propose an end-to-end method that refines the input point cloud and directly produces high-quality, artist-style meshes. At the core of our approach is a novel reformulation of 3D point cloud refinement as a 2D inpainting task, enabling the use of powerful generative models. Preliminary results on the ShapeNet dataset demonstrate the promise of our framework in producing clean, complete meshes.
Integrating Anatomical Priors into a Causal Diffusion Model
Sep 10, 2025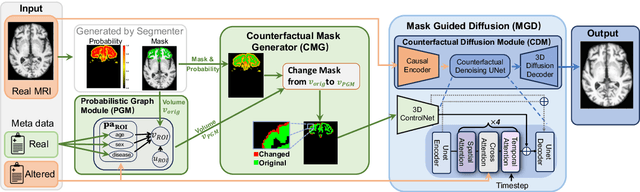
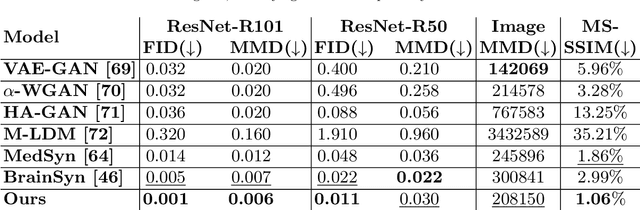
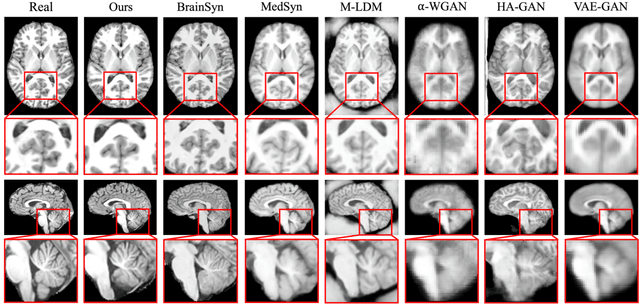
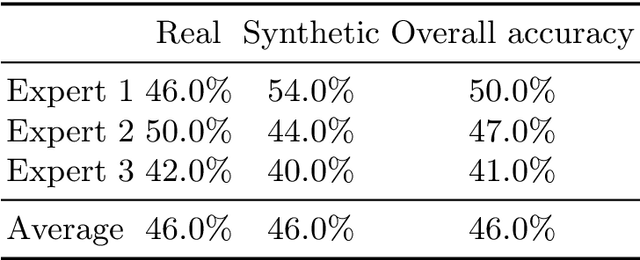
Abstract:3D brain MRI studies often examine subtle morphometric differences between cohorts that are hard to detect visually. Given the high cost of MRI acquisition, these studies could greatly benefit from image syntheses, particularly counterfactual image generation, as seen in other domains, such as computer vision. However, counterfactual models struggle to produce anatomically plausible MRIs due to the lack of explicit inductive biases to preserve fine-grained anatomical details. This shortcoming arises from the training of the models aiming to optimize for the overall appearance of the images (e.g., via cross-entropy) rather than preserving subtle, yet medically relevant, local variations across subjects. To preserve subtle variations, we propose to explicitly integrate anatomical constraints on a voxel-level as prior into a generative diffusion framework. Called Probabilistic Causal Graph Model (PCGM), the approach captures anatomical constraints via a probabilistic graph module and translates those constraints into spatial binary masks of regions where subtle variations occur. The masks (encoded by a 3D extension of ControlNet) constrain a novel counterfactual denoising UNet, whose encodings are then transferred into high-quality brain MRIs via our 3D diffusion decoder. Extensive experiments on multiple datasets demonstrate that PCGM generates structural brain MRIs of higher quality than several baseline approaches. Furthermore, we show for the first time that brain measurements extracted from counterfactuals (generated by PCGM) replicate the subtle effects of a disease on cortical brain regions previously reported in the neuroscience literature. This achievement is an important milestone in the use of synthetic MRIs in studies investigating subtle morphological differences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge