Medqa Usmle
Papers and Code
Capabilities of GPT-5 on Multimodal Medical Reasoning
Aug 13, 2025Recent advances in large language models (LLMs) have enabled general-purpose systems to perform increasingly complex domain-specific reasoning without extensive fine-tuning. In the medical domain, decision-making often requires integrating heterogeneous information sources, including patient narratives, structured data, and medical images. This study positions GPT-5 as a generalist multimodal reasoner for medical decision support and systematically evaluates its zero-shot chain-of-thought reasoning performance on both text-based question answering and visual question answering tasks under a unified protocol. We benchmark GPT-5, GPT-5-mini, GPT-5-nano, and GPT-4o-2024-11-20 against standardized splits of MedQA, MedXpertQA (text and multimodal), MMLU medical subsets, USMLE self-assessment exams, and VQA-RAD. Results show that GPT-5 consistently outperforms all baselines, achieving state-of-the-art accuracy across all QA benchmarks and delivering substantial gains in multimodal reasoning. On MedXpertQA MM, GPT-5 improves reasoning and understanding scores by +29.26% and +26.18% over GPT-4o, respectively, and surpasses pre-licensed human experts by +24.23% in reasoning and +29.40% in understanding. In contrast, GPT-4o remains below human expert performance in most dimensions. A representative case study demonstrates GPT-5's ability to integrate visual and textual cues into a coherent diagnostic reasoning chain, recommending appropriate high-stakes interventions. Our results show that, on these controlled multimodal reasoning benchmarks, GPT-5 moves from human-comparable to above human-expert performance. This improvement may substantially inform the design of future clinical decision-support systems.
WiNGPT-3.0 Technical Report
May 23, 2025



Current Large Language Models (LLMs) exhibit significant limitations, notably in structured, interpretable, and verifiable medical reasoning, alongside practical deployment challenges related to computational resources and data privacy. This report focused on the development of WiNGPT-3.0, the 32-billion parameter LLMs, engineered with the objective of enhancing its capacity for medical reasoning and exploring its potential for effective integration within healthcare IT infrastructures. The broader aim is to advance towards clinically applicable models. The approach involved a multi-stage training pipeline tailored for general, medical, and clinical reasoning. This pipeline incorporated supervised fine-tuning (SFT) and reinforcement learning (RL), leveraging curated Long Chain-of-Thought (CoT) datasets, auxiliary reward models, and an evidence-based diagnostic chain simulation. WiNGPT-3.0 demonstrated strong performance: specific model variants achieved scores of 66.6 on MedCalc and 87.1 on MedQA-USMLE. Furthermore, targeted training improved performance on a clinical reasoning task from a baseline score of 58.1 to 62.5. These findings suggest that reinforcement learning, even when applied with a limited dataset of only a few thousand examples, can enhance medical reasoning accuracy. Crucially, this demonstration of RL's efficacy with limited data and computation paves the way for more trustworthy and practically deployable LLMs within clinical workflows and health information infrastructures.
Disentangling Reasoning and Knowledge in Medical Large Language Models
May 16, 2025



Medical reasoning in large language models (LLMs) aims to emulate clinicians' diagnostic thinking, but current benchmarks such as MedQA-USMLE, MedMCQA, and PubMedQA often mix reasoning with factual recall. We address this by separating 11 biomedical QA benchmarks into reasoning- and knowledge-focused subsets using a PubMedBERT classifier that reaches 81 percent accuracy, comparable to human performance. Our analysis shows that only 32.8 percent of questions require complex reasoning. We evaluate biomedical models (HuatuoGPT-o1, MedReason, m1) and general-domain models (DeepSeek-R1, o4-mini, Qwen3), finding consistent gaps between knowledge and reasoning performance. For example, m1 scores 60.5 on knowledge but only 47.1 on reasoning. In adversarial tests where models are misled with incorrect initial reasoning, biomedical models degrade sharply, while larger or RL-trained general models show more robustness. To address this, we train BioMed-R1 using fine-tuning and reinforcement learning on reasoning-heavy examples. It achieves the strongest performance among similarly sized models. Further gains may come from incorporating clinical case reports and training with adversarial and backtracking scenarios.
Open-Medical-R1: How to Choose Data for RLVR Training at Medicine Domain
Apr 16, 2025



This paper explores optimal data selection strategies for Reinforcement Learning with Verified Rewards (RLVR) training in the medical domain. While RLVR has shown exceptional potential for enhancing reasoning capabilities in large language models, most prior implementations have focused on mathematics and logical puzzles, with limited exploration of domain-specific applications like medicine. We investigate four distinct data sampling strategies from MedQA-USMLE: random sampling (baseline), and filtering using Phi-4, Gemma-3-27b-it, and Gemma-3-12b-it models. Using Gemma-3-12b-it as our base model and implementing Group Relative Policy Optimization (GRPO), we evaluate performance across multiple benchmarks including MMLU, GSM8K, MMLU-Pro, and CMMLU. Our findings demonstrate that models trained on filtered data generally outperform those trained on randomly selected samples. Notably, training on self-filtered samples (using Gemma-3-12b-it for filtering) achieved superior performance in medical domains but showed reduced robustness across different benchmarks, while filtering with larger models from the same series yielded better overall robustness. These results provide valuable insights into effective data organization strategies for RLVR in specialized domains and highlight the importance of thoughtful data selection in achieving optimal performance. You can access our repository (https://github.com/Qsingle/open-medical-r1) to get the codes.
AfriMed-QA: A Pan-African, Multi-Specialty, Medical Question-Answering Benchmark Dataset
Nov 23, 2024

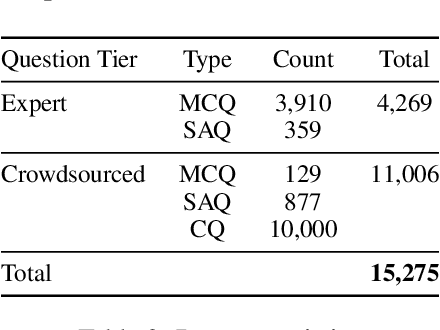

Recent advancements in large language model(LLM) performance on medical multiple choice question (MCQ) benchmarks have stimulated interest from healthcare providers and patients globally. Particularly in low-and middle-income countries (LMICs) facing acute physician shortages and lack of specialists, LLMs offer a potentially scalable pathway to enhance healthcare access and reduce costs. However, their effectiveness in the Global South, especially across the African continent, remains to be established. In this work, we introduce AfriMed-QA, the first large scale Pan-African English multi-specialty medical Question-Answering (QA) dataset, 15,000 questions (open and closed-ended) sourced from over 60 medical schools across 16 countries, covering 32 medical specialties. We further evaluate 30 LLMs across multiple axes including correctness and demographic bias. Our findings show significant performance variation across specialties and geographies, MCQ performance clearly lags USMLE (MedQA). We find that biomedical LLMs underperform general models and smaller edge-friendly LLMs struggle to achieve a passing score. Interestingly, human evaluations show a consistent consumer preference for LLM answers and explanations when compared with clinician answers.
MedMobile: A mobile-sized language model with expert-level clinical capabilities
Oct 11, 2024


Language models (LMs) have demonstrated expert-level reasoning and recall abilities in medicine. However, computational costs and privacy concerns are mounting barriers to wide-scale implementation. We introduce a parsimonious adaptation of phi-3-mini, MedMobile, a 3.8 billion parameter LM capable of running on a mobile device, for medical applications. We demonstrate that MedMobile scores 75.7% on the MedQA (USMLE), surpassing the passing mark for physicians (~60%), and approaching the scores of models 100 times its size. We subsequently perform a careful set of ablations, and demonstrate that chain of thought, ensembling, and fine-tuning lead to the greatest performance gains, while unexpectedly retrieval augmented generation fails to demonstrate significant improvements
Capabilities of Gemini Models in Medicine
May 01, 2024



Excellence in a wide variety of medical applications poses considerable challenges for AI, requiring advanced reasoning, access to up-to-date medical knowledge and understanding of complex multimodal data. Gemini models, with strong general capabilities in multimodal and long-context reasoning, offer exciting possibilities in medicine. Building on these core strengths of Gemini, we introduce Med-Gemini, a family of highly capable multimodal models that are specialized in medicine with the ability to seamlessly use web search, and that can be efficiently tailored to novel modalities using custom encoders. We evaluate Med-Gemini on 14 medical benchmarks, establishing new state-of-the-art (SoTA) performance on 10 of them, and surpass the GPT-4 model family on every benchmark where a direct comparison is viable, often by a wide margin. On the popular MedQA (USMLE) benchmark, our best-performing Med-Gemini model achieves SoTA performance of 91.1% accuracy, using a novel uncertainty-guided search strategy. On 7 multimodal benchmarks including NEJM Image Challenges and MMMU (health & medicine), Med-Gemini improves over GPT-4V by an average relative margin of 44.5%. We demonstrate the effectiveness of Med-Gemini's long-context capabilities through SoTA performance on a needle-in-a-haystack retrieval task from long de-identified health records and medical video question answering, surpassing prior bespoke methods using only in-context learning. Finally, Med-Gemini's performance suggests real-world utility by surpassing human experts on tasks such as medical text summarization, alongside demonstrations of promising potential for multimodal medical dialogue, medical research and education. Taken together, our results offer compelling evidence for Med-Gemini's potential, although further rigorous evaluation will be crucial before real-world deployment in this safety-critical domain.
Few shot chain-of-thought driven reasoning to prompt LLMs for open ended medical question answering
Mar 07, 2024



Large Language models (LLMs) have demonstrated significant potential in transforming healthcare by automating tasks such as clinical documentation, information retrieval, and decision support. In this aspect, carefully engineered prompts have emerged as a powerful tool for using LLMs for medical scenarios, e.g., patient clinical scenarios. In this paper, we propose a modified version of the MedQA-USMLE dataset, which is subjective, to mimic real-life clinical scenarios. We explore the Chain of Thought (CoT) reasoning based on subjective response generation for the modified MedQA-USMLE dataset with appropriate LM-driven forward reasoning for correct responses to the medical questions. Keeping in mind the importance of response verification in the medical setting, we utilize a reward training mechanism whereby the language model also provides an appropriate verified response for a particular response to a clinical question. In this regard, we also include human-in-the-loop for different evaluation aspects. We develop better in-contrast learning strategies by modifying the 5-shot-codex-CoT-prompt from arXiv:2207.08143 for the subjective MedQA dataset and developing our incremental-reasoning prompt. Our evaluations show that the incremental reasoning prompt performs better than the modified codex prompt in certain scenarios. We also show that greedy decoding with the incremental reasoning method performs better than other strategies, such as prompt chaining and eliminative reasoning.
To Generate or to Retrieve? On the Effectiveness of Artificial Contexts for Medical Open-Domain Question Answering
Mar 04, 2024



Medical open-domain question answering demands substantial access to specialized knowledge. Recent efforts have sought to decouple knowledge from model parameters, counteracting architectural scaling and allowing for training on common low-resource hardware. The retrieve-then-read paradigm has become ubiquitous, with model predictions grounded on relevant knowledge pieces from external repositories such as PubMed, textbooks, and UMLS. An alternative path, still under-explored but made possible by the advent of domain-specific large language models, entails constructing artificial contexts through prompting. As a result, "to generate or to retrieve" is the modern equivalent of Hamlet's dilemma. This paper presents MedGENIE, the first generate-then-read framework for multiple-choice question answering in medicine. We conduct extensive experiments on MedQA-USMLE, MedMCQA, and MMLU, incorporating a practical perspective by assuming a maximum of 24GB VRAM. MedGENIE sets a new state-of-the-art (SOTA) in the open-book setting of each testbed, even allowing a small-scale reader to outcompete zero-shot closed-book 175B baselines while using up to 706$\times$ fewer parameters. Overall, our findings reveal that generated passages are more effective than retrieved counterparts in attaining higher accuracy.
Improving Retrieval-Augmented Generation in Medicine with Iterative Follow-up Questions
Aug 01, 2024



The emergent abilities of large language models (LLMs) have demonstrated great potential in solving medical questions. They can possess considerable medical knowledge, but may still hallucinate and are inflexible in the knowledge updates. While Retrieval-Augmented Generation (RAG) has been proposed to enhance the medical question-answering capabilities of LLMs with external knowledge bases, it may still fail in complex cases where multiple rounds of information-seeking are required. To address such an issue, we propose iterative RAG for medicine (i-MedRAG), where LLMs can iteratively ask follow-up queries based on previous information-seeking attempts. In each iteration of i-MedRAG, the follow-up queries will be answered by a vanilla RAG system and they will be further used to guide the query generation in the next iteration. Our experiments show the improved performance of various LLMs brought by i-MedRAG compared with vanilla RAG on complex questions from clinical vignettes in the United States Medical Licensing Examination (USMLE), as well as various knowledge tests in the Massive Multitask Language Understanding (MMLU) dataset. Notably, our zero-shot i-MedRAG outperforms all existing prompt engineering and fine-tuning methods on GPT-3.5, achieving an accuracy of 69.68\% on the MedQA dataset. In addition, we characterize the scaling properties of i-MedRAG with different iterations of follow-up queries and different numbers of queries per iteration. Our case studies show that i-MedRAG can flexibly ask follow-up queries to form reasoning chains, providing an in-depth analysis of medical questions. To the best of our knowledge, this is the first-of-its-kind study on incorporating follow-up queries into medical RAG.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge