Yan Hu
A Federated and Parameter-Efficient Framework for Large Language Model Training in Medicine
Jan 29, 2026Abstract:Large language models (LLMs) have demonstrated strong performance on medical benchmarks, including question answering and diagnosis. To enable their use in clinical settings, LLMs are typically further adapted through continued pretraining or post-training using clinical data. However, most medical LLMs are trained on data from a single institution, which faces limitations in generalizability and safety in heterogeneous systems. Federated learning (FL) is a promising solution for enabling collaborative model development across healthcare institutions. Yet applying FL to LLMs in medicine remains fundamentally limited. First, conventional FL requires transmitting the full model during each communication round, which becomes impractical for multi-billion-parameter LLMs given the limited computational resources. Second, many FL algorithms implicitly assume data homogeneity, whereas real-world clinical data are highly heterogeneous across patients, diseases, and institutional practices. We introduce the model-agnostic and parameter-efficient federated learning framework for adapting LLMs to medical applications. Fed-MedLoRA transmits only low-rank adapter parameters, reducing communication and computation overhead, while Fed-MedLoRA+ further incorporates adaptive, data-aware aggregation to improve convergence under cross-site heterogeneity. We apply the framework to clinical information extraction (IE), which transforms patient narratives into structured medical entities and relations. Accuracy was assessed across five patient cohorts through comparisons with BERT models, and LLaMA-3 and DeepSeek-R1, GPT-4o models. Evaluation settings included (1) in-domain training and testing, (2) external validation on independent cohorts, and (3) a low-resource new-site adaptation scenario using real-world clinical notes from the Yale New Haven Health System.
Can Multimodal LLMs See Materials Clearly? A Multimodal Benchmark on Materials Characterization
Sep 11, 2025



Abstract:Materials characterization is fundamental to acquiring materials information, revealing the processing-microstructure-property relationships that guide material design and optimization. While multimodal large language models (MLLMs) have recently shown promise in generative and predictive tasks within materials science, their capacity to understand real-world characterization imaging data remains underexplored. To bridge this gap, we present MatCha, the first benchmark for materials characterization image understanding, comprising 1,500 questions that demand expert-level domain expertise. MatCha encompasses four key stages of materials research comprising 21 distinct tasks, each designed to reflect authentic challenges faced by materials scientists. Our evaluation of state-of-the-art MLLMs on MatCha reveals a significant performance gap compared to human experts. These models exhibit degradation when addressing questions requiring higher-level expertise and sophisticated visual perception. Simple few-shot and chain-of-thought prompting struggle to alleviate these limitations. These findings highlight that existing MLLMs still exhibit limited adaptability to real-world materials characterization scenarios. We hope MatCha will facilitate future research in areas such as new material discovery and autonomous scientific agents. MatCha is available at https://github.com/FreedomIntelligence/MatCha.
Memorization in Large Language Models in Medicine: Prevalence, Characteristics, and Implications
Sep 10, 2025Abstract:Large Language Models (LLMs) have demonstrated significant potential in medicine. To date, LLMs have been widely applied to tasks such as diagnostic assistance, medical question answering, and clinical information synthesis. However, a key open question remains: to what extent do LLMs memorize medical training data. In this study, we present the first comprehensive evaluation of memorization of LLMs in medicine, assessing its prevalence (how frequently it occurs), characteristics (what is memorized), volume (how much content is memorized), and potential downstream impacts (how memorization may affect medical applications). We systematically analyze common adaptation scenarios: (1) continued pretraining on medical corpora, (2) fine-tuning on standard medical benchmarks, and (3) fine-tuning on real-world clinical data, including over 13,000 unique inpatient records from Yale New Haven Health System. The results demonstrate that memorization is prevalent across all adaptation scenarios and significantly higher than reported in the general domain. Memorization affects both the development and adoption of LLMs in medicine and can be categorized into three types: beneficial (e.g., accurate recall of clinical guidelines and biomedical references), uninformative (e.g., repeated disclaimers or templated medical document language), and harmful (e.g., regeneration of dataset-specific or sensitive clinical content). Based on these findings, we offer practical recommendations to facilitate beneficial memorization that enhances domain-specific reasoning and factual accuracy, minimize uninformative memorization to promote deeper learning beyond surface-level patterns, and mitigate harmful memorization to prevent the leakage of sensitive or identifiable patient information.
Towards Assessing Medical Ethics from Knowledge to Practice
Aug 07, 2025Abstract:The integration of large language models into healthcare necessitates a rigorous evaluation of their ethical reasoning, an area current benchmarks often overlook. We introduce PrinciplismQA, a comprehensive benchmark with 3,648 questions designed to systematically assess LLMs' alignment with core medical ethics. Grounded in Principlism, our benchmark features a high-quality dataset. This includes multiple-choice questions curated from authoritative textbooks and open-ended questions sourced from authoritative medical ethics case study literature, all validated by medical experts. Our experiments reveal a significant gap between models' ethical knowledge and their practical application, especially in dynamically applying ethical principles to real-world scenarios. Most LLMs struggle with dilemmas concerning Beneficence, often over-emphasizing other principles. Frontier closed-source models, driven by strong general capabilities, currently lead the benchmark. Notably, medical domain fine-tuning can enhance models' overall ethical competence, but further progress requires better alignment with medical ethical knowledge. PrinciplismQA offers a scalable framework to diagnose these specific ethical weaknesses, paving the way for more balanced and responsible medical AI.
Multi-Label Classification with Generative AI Models in Healthcare: A Case Study of Suicidality and Risk Factors
Jul 22, 2025



Abstract:Suicide remains a pressing global health crisis, with over 720,000 deaths annually and millions more affected by suicide ideation (SI) and suicide attempts (SA). Early identification of suicidality-related factors (SrFs), including SI, SA, exposure to suicide (ES), and non-suicidal self-injury (NSSI), is critical for timely intervention. While prior studies have applied AI to detect SrFs in clinical notes, most treat suicidality as a binary classification task, overlooking the complexity of cooccurring risk factors. This study explores the use of generative large language models (LLMs), specifically GPT-3.5 and GPT-4.5, for multi-label classification (MLC) of SrFs from psychiatric electronic health records (EHRs). We present a novel end to end generative MLC pipeline and introduce advanced evaluation methods, including label set level metrics and a multilabel confusion matrix for error analysis. Finetuned GPT-3.5 achieved top performance with 0.94 partial match accuracy and 0.91 F1 score, while GPT-4.5 with guided prompting showed superior performance across label sets, including rare or minority label sets, indicating a more balanced and robust performance. Our findings reveal systematic error patterns, such as the conflation of SI and SA, and highlight the models tendency toward cautious over labeling. This work not only demonstrates the feasibility of using generative AI for complex clinical classification tasks but also provides a blueprint for structuring unstructured EHR data to support large scale clinical research and evidence based medicine.
Model Unlearning via Sparse Autoencoder Subspace Guided Projections
May 30, 2025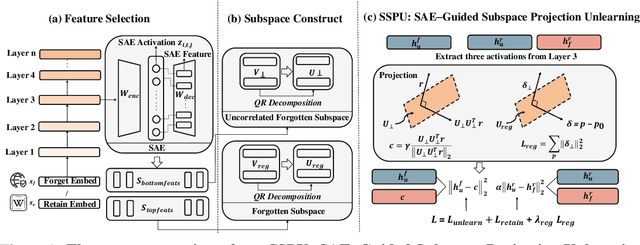



Abstract:Large language models (LLMs) store vast amounts of information, making them powerful yet raising privacy and safety concerns when selective knowledge removal is required. Existing unlearning strategies, ranging from gradient-based fine-tuning and model editing to sparse autoencoder (SAE) steering, either lack interpretability or fail to provide a robust defense against adversarial prompts. We propose SAE-Guided Subspace Projection Unlearning (SSPU), a novel framework that leverages SAE features to drive targeted updates in the model's parameter space, enabling precise, interpretable, and robust unlearning. SSPU's three-stage pipeline performs data-driven layer and feature selection, subspace construction via QR decomposition, and constrained optimization that controls activations into an "irrelevant" subspace while preserving retained knowledge. Overall, we use SAE features to construct a subspace that supervises unlearning, refining the loss and adding a regularization term to guide interpretable parameter updates. In experiments on the WMDP-Cyber forget set and three utility benchmarks (MMLU, TruthfulQA, GSM8K), SSPU reduces harmful knowledge accuracy by 3.22% compared to the strongest baseline. It also improves adversarial robustness, lowering malicious accuracy under jailbreak prompts compared to baselines. Our findings expose the limitations of prior unlearning methods and demonstrate how interpretable subspace-guided optimization can achieve robust, controllable model behavior.
RADAR: Enhancing Radiology Report Generation with Supplementary Knowledge Injection
May 20, 2025Abstract:Large language models (LLMs) have demonstrated remarkable capabilities in various domains, including radiology report generation. Previous approaches have attempted to utilize multimodal LLMs for this task, enhancing their performance through the integration of domain-specific knowledge retrieval. However, these approaches often overlook the knowledge already embedded within the LLMs, leading to redundant information integration and inefficient utilization of learned representations. To address this limitation, we propose RADAR, a framework for enhancing radiology report generation with supplementary knowledge injection. RADAR improves report generation by systematically leveraging both the internal knowledge of an LLM and externally retrieved information. Specifically, it first extracts the model's acquired knowledge that aligns with expert image-based classification outputs. It then retrieves relevant supplementary knowledge to further enrich this information. Finally, by aggregating both sources, RADAR generates more accurate and informative radiology reports. Extensive experiments on MIMIC-CXR, CheXpert-Plus, and IU X-ray demonstrate that our model outperforms state-of-the-art LLMs in both language quality and clinical accuracy
Open-Medical-R1: How to Choose Data for RLVR Training at Medicine Domain
Apr 16, 2025



Abstract:This paper explores optimal data selection strategies for Reinforcement Learning with Verified Rewards (RLVR) training in the medical domain. While RLVR has shown exceptional potential for enhancing reasoning capabilities in large language models, most prior implementations have focused on mathematics and logical puzzles, with limited exploration of domain-specific applications like medicine. We investigate four distinct data sampling strategies from MedQA-USMLE: random sampling (baseline), and filtering using Phi-4, Gemma-3-27b-it, and Gemma-3-12b-it models. Using Gemma-3-12b-it as our base model and implementing Group Relative Policy Optimization (GRPO), we evaluate performance across multiple benchmarks including MMLU, GSM8K, MMLU-Pro, and CMMLU. Our findings demonstrate that models trained on filtered data generally outperform those trained on randomly selected samples. Notably, training on self-filtered samples (using Gemma-3-12b-it for filtering) achieved superior performance in medical domains but showed reduced robustness across different benchmarks, while filtering with larger models from the same series yielded better overall robustness. These results provide valuable insights into effective data organization strategies for RLVR in specialized domains and highlight the importance of thoughtful data selection in achieving optimal performance. You can access our repository (https://github.com/Qsingle/open-medical-r1) to get the codes.
Adaptive Wavelet Filters as Practical Texture Feature Amplifiers for Parkinson's Disease Screening in OCT
Mar 25, 2025Abstract:Parkinson's disease (PD) is a prevalent neurodegenerative disorder globally. The eye's retina is an extension of the brain and has great potential in PD screening. Recent studies have suggested that texture features extracted from retinal layers can be adopted as biomarkers for PD diagnosis under optical coherence tomography (OCT) images. Frequency domain learning techniques can enhance the feature representations of deep neural networks (DNNs) by decomposing frequency components involving rich texture features. Additionally, previous works have not exploited texture features for automated PD screening in OCT. Motivated by the above analysis, we propose a novel Adaptive Wavelet Filter (AWF) that serves as the Practical Texture Feature Amplifier to fully leverage the merits of texture features to boost the PD screening performance of DNNs with the aid of frequency domain learning. Specifically, AWF first enhances texture feature representation diversities via channel mixer, then emphasizes informative texture feature representations with the well-designed adaptive wavelet filtering token mixer. By combining the AWFs with the DNN stem, AWFNet is constructed for automated PD screening. Additionally, we introduce a novel Balanced Confidence (BC) Loss by mining the potential of sample-wise predicted probabilities of all classes and class frequency prior, to further boost the PD screening performance and trustworthiness of AWFNet. The extensive experiments manifest the superiority of our AWFNet and BC over state-of-the-art methods in terms of PD screening performance and trustworthiness.
Generalizable and Explainable Deep Learning for Medical Image Computing: An Overview
Mar 11, 2025



Abstract:Objective. This paper presents an overview of generalizable and explainable artificial intelligence (XAI) in deep learning (DL) for medical imaging, aimed at addressing the urgent need for transparency and explainability in clinical applications. Methodology. We propose to use four CNNs in three medical datasets (brain tumor, skin cancer, and chest x-ray) for medical image classification tasks. In addition, we perform paired t-tests to show the significance of the differences observed between different methods. Furthermore, we propose to combine ResNet50 with five common XAI techniques to obtain explainable results for model prediction, aiming at improving model transparency. We also involve a quantitative metric (confidence increase) to evaluate the usefulness of XAI techniques. Key findings. The experimental results indicate that ResNet50 can achieve feasible accuracy and F1 score in all datasets (e.g., 86.31\% accuracy in skin cancer). Furthermore, the findings show that while certain XAI methods, such as XgradCAM, effectively highlight relevant abnormal regions in medical images, others, like EigenGradCAM, may perform less effectively in specific scenarios. In addition, XgradCAM indicates higher confidence increase (e.g., 0.12 in glioma tumor) compared to GradCAM++ (0.09) and LayerCAM (0.08). Implications. Based on the experimental results and recent advancements, we outline future research directions to enhance the robustness and generalizability of DL models in the field of biomedical imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge