Ellery Wulczyn
PathAlign: A vision-language model for whole slide images in histopathology
Jun 27, 2024Abstract:Microscopic interpretation of histopathology images underlies many important diagnostic and treatment decisions. While advances in vision-language modeling raise new opportunities for analysis of such images, the gigapixel-scale size of whole slide images (WSIs) introduces unique challenges. Additionally, pathology reports simultaneously highlight key findings from small regions while also aggregating interpretation across multiple slides, often making it difficult to create robust image-text pairs. As such, pathology reports remain a largely untapped source of supervision in computational pathology, with most efforts relying on region-of-interest annotations or self-supervision at the patch-level. In this work, we develop a vision-language model based on the BLIP-2 framework using WSIs paired with curated text from pathology reports. This enables applications utilizing a shared image-text embedding space, such as text or image retrieval for finding cases of interest, as well as integration of the WSI encoder with a frozen large language model (LLM) for WSI-based generative text capabilities such as report generation or AI-in-the-loop interactions. We utilize a de-identified dataset of over 350,000 WSIs and diagnostic text pairs, spanning a wide range of diagnoses, procedure types, and tissue types. We present pathologist evaluation of text generation and text retrieval using WSI embeddings, as well as results for WSI classification and workflow prioritization (slide-level triaging). Model-generated text for WSIs was rated by pathologists as accurate, without clinically significant error or omission, for 78% of WSIs on average. This work demonstrates exciting potential capabilities for language-aligned WSI embeddings.
Advancing Multimodal Medical Capabilities of Gemini
May 06, 2024



Abstract:Many clinical tasks require an understanding of specialized data, such as medical images and genomics, which is not typically found in general-purpose large multimodal models. Building upon Gemini's multimodal models, we develop several models within the new Med-Gemini family that inherit core capabilities of Gemini and are optimized for medical use via fine-tuning with 2D and 3D radiology, histopathology, ophthalmology, dermatology and genomic data. Med-Gemini-2D sets a new standard for AI-based chest X-ray (CXR) report generation based on expert evaluation, exceeding previous best results across two separate datasets by an absolute margin of 1% and 12%, where 57% and 96% of AI reports on normal cases, and 43% and 65% on abnormal cases, are evaluated as "equivalent or better" than the original radiologists' reports. We demonstrate the first ever large multimodal model-based report generation for 3D computed tomography (CT) volumes using Med-Gemini-3D, with 53% of AI reports considered clinically acceptable, although additional research is needed to meet expert radiologist reporting quality. Beyond report generation, Med-Gemini-2D surpasses the previous best performance in CXR visual question answering (VQA) and performs well in CXR classification and radiology VQA, exceeding SoTA or baselines on 17 of 20 tasks. In histopathology, ophthalmology, and dermatology image classification, Med-Gemini-2D surpasses baselines across 18 out of 20 tasks and approaches task-specific model performance. Beyond imaging, Med-Gemini-Polygenic outperforms the standard linear polygenic risk score-based approach for disease risk prediction and generalizes to genetically correlated diseases for which it has never been trained. Although further development and evaluation are necessary in the safety-critical medical domain, our results highlight the potential of Med-Gemini across a wide range of medical tasks.
Capabilities of Gemini Models in Medicine
May 01, 2024



Abstract:Excellence in a wide variety of medical applications poses considerable challenges for AI, requiring advanced reasoning, access to up-to-date medical knowledge and understanding of complex multimodal data. Gemini models, with strong general capabilities in multimodal and long-context reasoning, offer exciting possibilities in medicine. Building on these core strengths of Gemini, we introduce Med-Gemini, a family of highly capable multimodal models that are specialized in medicine with the ability to seamlessly use web search, and that can be efficiently tailored to novel modalities using custom encoders. We evaluate Med-Gemini on 14 medical benchmarks, establishing new state-of-the-art (SoTA) performance on 10 of them, and surpass the GPT-4 model family on every benchmark where a direct comparison is viable, often by a wide margin. On the popular MedQA (USMLE) benchmark, our best-performing Med-Gemini model achieves SoTA performance of 91.1% accuracy, using a novel uncertainty-guided search strategy. On 7 multimodal benchmarks including NEJM Image Challenges and MMMU (health & medicine), Med-Gemini improves over GPT-4V by an average relative margin of 44.5%. We demonstrate the effectiveness of Med-Gemini's long-context capabilities through SoTA performance on a needle-in-a-haystack retrieval task from long de-identified health records and medical video question answering, surpassing prior bespoke methods using only in-context learning. Finally, Med-Gemini's performance suggests real-world utility by surpassing human experts on tasks such as medical text summarization, alongside demonstrations of promising potential for multimodal medical dialogue, medical research and education. Taken together, our results offer compelling evidence for Med-Gemini's potential, although further rigorous evaluation will be crucial before real-world deployment in this safety-critical domain.
Domain-specific optimization and diverse evaluation of self-supervised models for histopathology
Oct 20, 2023



Abstract:Task-specific deep learning models in histopathology offer promising opportunities for improving diagnosis, clinical research, and precision medicine. However, development of such models is often limited by availability of high-quality data. Foundation models in histopathology that learn general representations across a wide range of tissue types, diagnoses, and magnifications offer the potential to reduce the data, compute, and technical expertise necessary to develop task-specific deep learning models with the required level of model performance. In this work, we describe the development and evaluation of foundation models for histopathology via self-supervised learning (SSL). We first establish a diverse set of benchmark tasks involving 17 unique tissue types and 12 unique cancer types and spanning different optimal magnifications and task types. Next, we use this benchmark to explore and evaluate histopathology-specific SSL methods followed by further evaluation on held out patch-level and weakly supervised tasks. We found that standard SSL methods thoughtfully applied to histopathology images are performant across our benchmark tasks and that domain-specific methodological improvements can further increase performance. Our findings reinforce the value of using domain-specific SSL methods in pathology, and establish a set of high quality foundation models to enable further research across diverse applications.
Towards Expert-Level Medical Question Answering with Large Language Models
May 16, 2023



Abstract:Recent artificial intelligence (AI) systems have reached milestones in "grand challenges" ranging from Go to protein-folding. The capability to retrieve medical knowledge, reason over it, and answer medical questions comparably to physicians has long been viewed as one such grand challenge. Large language models (LLMs) have catalyzed significant progress in medical question answering; Med-PaLM was the first model to exceed a "passing" score in US Medical Licensing Examination (USMLE) style questions with a score of 67.2% on the MedQA dataset. However, this and other prior work suggested significant room for improvement, especially when models' answers were compared to clinicians' answers. Here we present Med-PaLM 2, which bridges these gaps by leveraging a combination of base LLM improvements (PaLM 2), medical domain finetuning, and prompting strategies including a novel ensemble refinement approach. Med-PaLM 2 scored up to 86.5% on the MedQA dataset, improving upon Med-PaLM by over 19% and setting a new state-of-the-art. We also observed performance approaching or exceeding state-of-the-art across MedMCQA, PubMedQA, and MMLU clinical topics datasets. We performed detailed human evaluations on long-form questions along multiple axes relevant to clinical applications. In pairwise comparative ranking of 1066 consumer medical questions, physicians preferred Med-PaLM 2 answers to those produced by physicians on eight of nine axes pertaining to clinical utility (p < 0.001). We also observed significant improvements compared to Med-PaLM on every evaluation axis (p < 0.001) on newly introduced datasets of 240 long-form "adversarial" questions to probe LLM limitations. While further studies are necessary to validate the efficacy of these models in real-world settings, these results highlight rapid progress towards physician-level performance in medical question answering.
Robust and Efficient Medical Imaging with Self-Supervision
May 19, 2022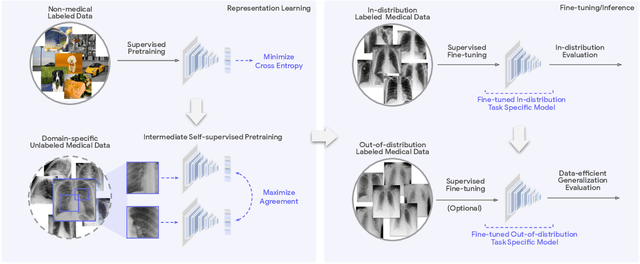
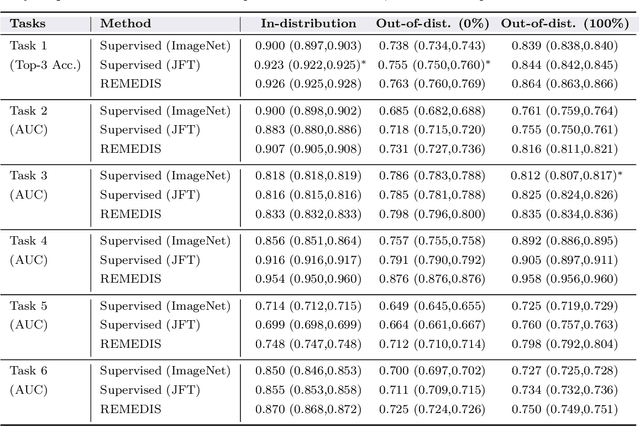
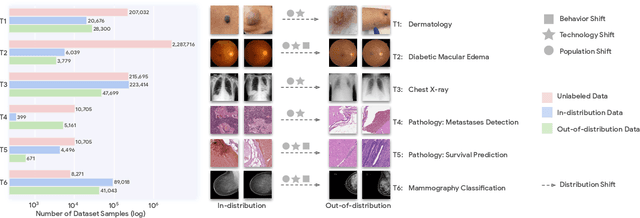
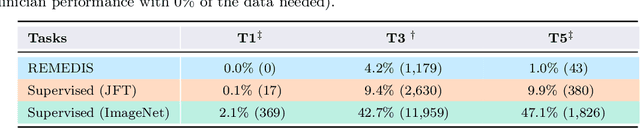
Abstract:Recent progress in Medical Artificial Intelligence (AI) has delivered systems that can reach clinical expert level performance. However, such systems tend to demonstrate sub-optimal "out-of-distribution" performance when evaluated in clinical settings different from the training environment. A common mitigation strategy is to develop separate systems for each clinical setting using site-specific data [1]. However, this quickly becomes impractical as medical data is time-consuming to acquire and expensive to annotate [2]. Thus, the problem of "data-efficient generalization" presents an ongoing difficulty for Medical AI development. Although progress in representation learning shows promise, their benefits have not been rigorously studied, specifically for out-of-distribution settings. To meet these challenges, we present REMEDIS, a unified representation learning strategy to improve robustness and data-efficiency of medical imaging AI. REMEDIS uses a generic combination of large-scale supervised transfer learning with self-supervised learning and requires little task-specific customization. We study a diverse range of medical imaging tasks and simulate three realistic application scenarios using retrospective data. REMEDIS exhibits significantly improved in-distribution performance with up to 11.5% relative improvement in diagnostic accuracy over a strong supervised baseline. More importantly, our strategy leads to strong data-efficient generalization of medical imaging AI, matching strong supervised baselines using between 1% to 33% of retraining data across tasks. These results suggest that REMEDIS can significantly accelerate the life-cycle of medical imaging AI development thereby presenting an important step forward for medical imaging AI to deliver broad impact.
Predicting Prostate Cancer-Specific Mortality with A.I.-based Gleason Grading
Nov 25, 2020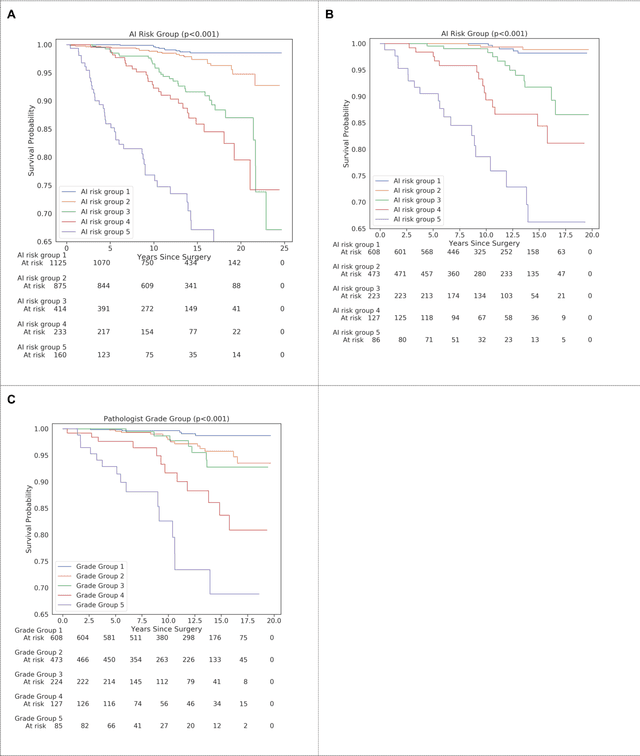
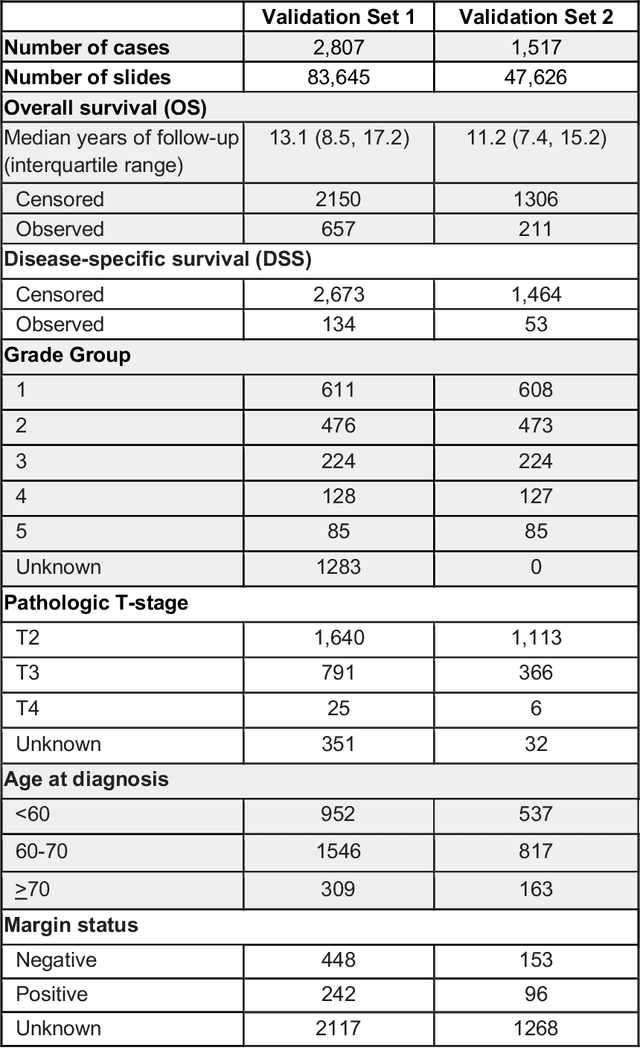
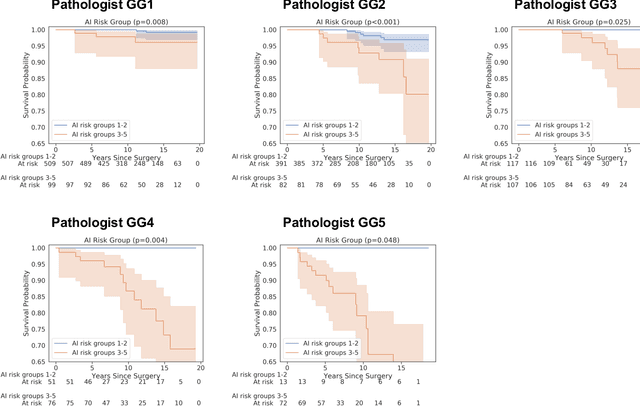
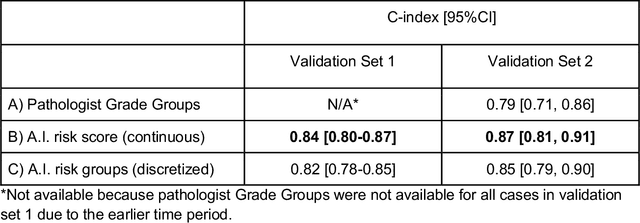
Abstract:Gleason grading of prostate cancer is an important prognostic factor but suffers from poor reproducibility, particularly among non-subspecialist pathologists. Although artificial intelligence (A.I.) tools have demonstrated Gleason grading on-par with expert pathologists, it remains an open question whether A.I. grading translates to better prognostication. In this study, we developed a system to predict prostate-cancer specific mortality via A.I.-based Gleason grading and subsequently evaluated its ability to risk-stratify patients on an independent retrospective cohort of 2,807 prostatectomy cases from a single European center with 5-25 years of follow-up (median: 13, interquartile range 9-17). The A.I.'s risk scores produced a C-index of 0.84 (95%CI 0.80-0.87) for prostate cancer-specific mortality. Upon discretizing these risk scores into risk groups analogous to pathologist Grade Groups (GG), the A.I. had a C-index of 0.82 (95%CI 0.78-0.85). On the subset of cases with a GG in the original pathology report (n=1,517), the A.I.'s C-indices were 0.87 and 0.85 for continuous and discrete grading, respectively, compared to 0.79 (95%CI 0.71-0.86) for GG obtained from the reports. These represent improvements of 0.08 (95%CI 0.01-0.15) and 0.07 (95%CI 0.00-0.14) respectively. Our results suggest that A.I.-based Gleason grading can lead to effective risk-stratification and warrants further evaluation for improving disease management.
Interpretable Survival Prediction for Colorectal Cancer using Deep Learning
Nov 17, 2020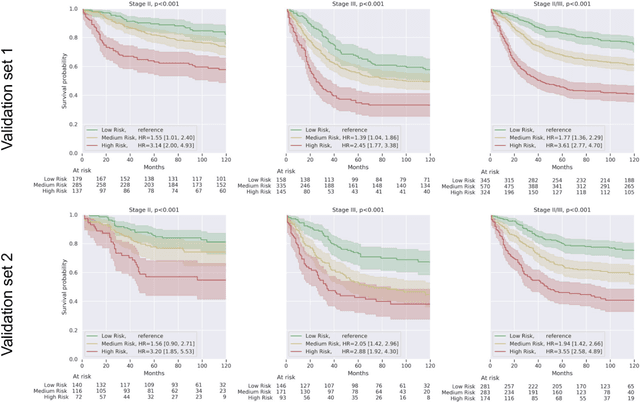
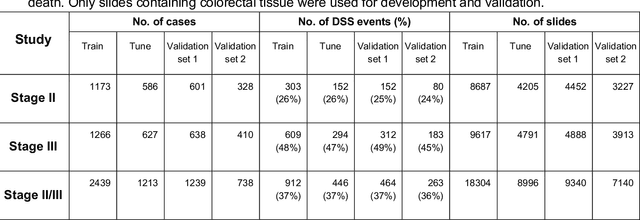
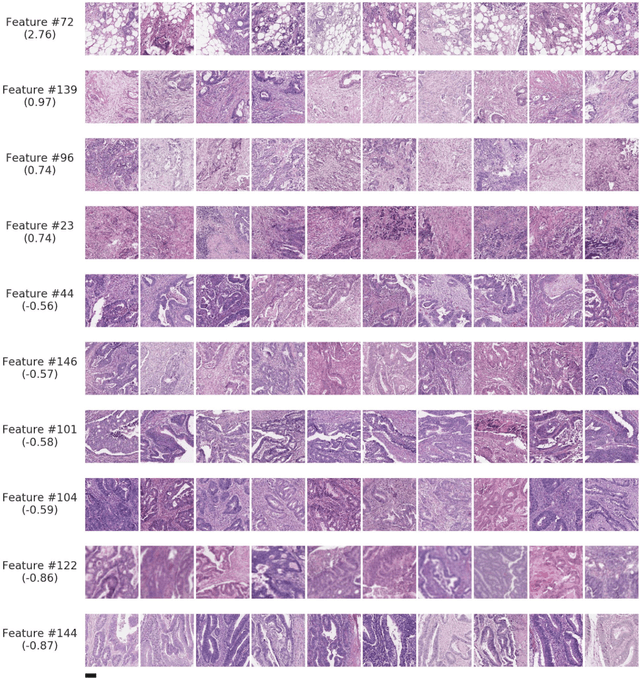
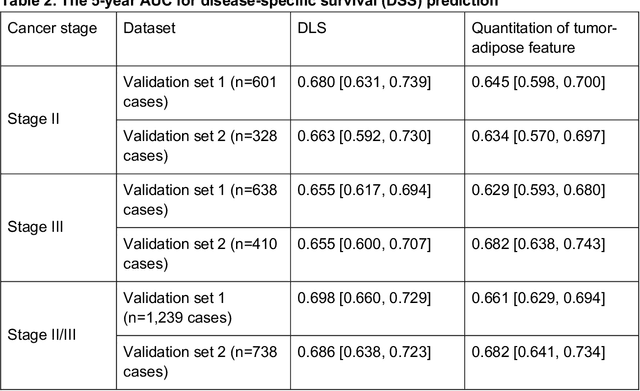
Abstract:Deriving interpretable prognostic features from deep-learning-based prognostic histopathology models remains a challenge. In this study, we developed a deep learning system (DLS) for predicting disease specific survival for stage II and III colorectal cancer using 3,652 cases (27,300 slides). When evaluated on two validation datasets containing 1,239 cases (9,340 slides) and 738 cases (7,140 slides) respectively, the DLS achieved a 5-year disease-specific survival AUC of 0.70 (95%CI 0.66-0.73) and 0.69 (95%CI 0.64-0.72), and added significant predictive value to a set of 9 clinicopathologic features. To interpret the DLS, we explored the ability of different human-interpretable features to explain the variance in DLS scores. We observed that clinicopathologic features such as T-category, N-category, and grade explained a small fraction of the variance in DLS scores (R2=18% in both validation sets). Next, we generated human-interpretable histologic features by clustering embeddings from a deep-learning based image-similarity model and showed that they explain the majority of the variance (R2 of 73% to 80%). Furthermore, the clustering-derived feature most strongly associated with high DLS scores was also highly prognostic in isolation. With a distinct visual appearance (poorly differentiated tumor cell clusters adjacent to adipose tissue), this feature was identified by annotators with 87.0-95.5% accuracy. Our approach can be used to explain predictions from a prognostic deep learning model and uncover potentially-novel prognostic features that can be reliably identified by people for future validation studies.
Deep learning-based survival prediction for multiple cancer types using histopathology images
Dec 16, 2019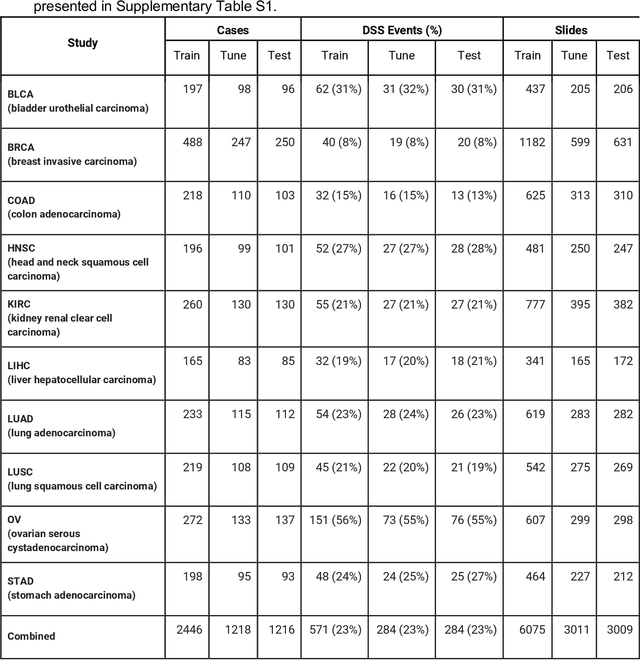
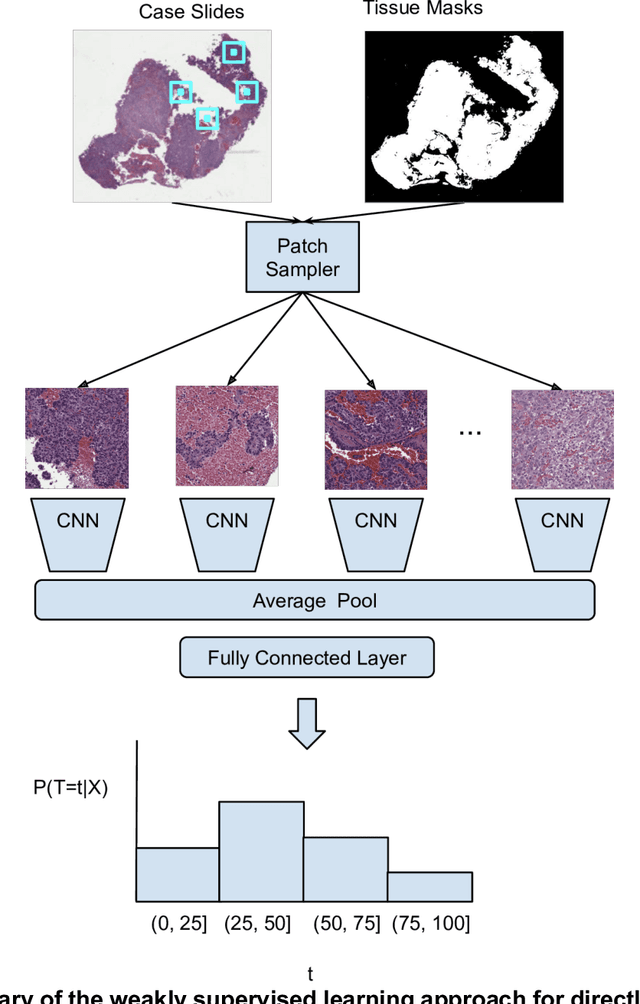
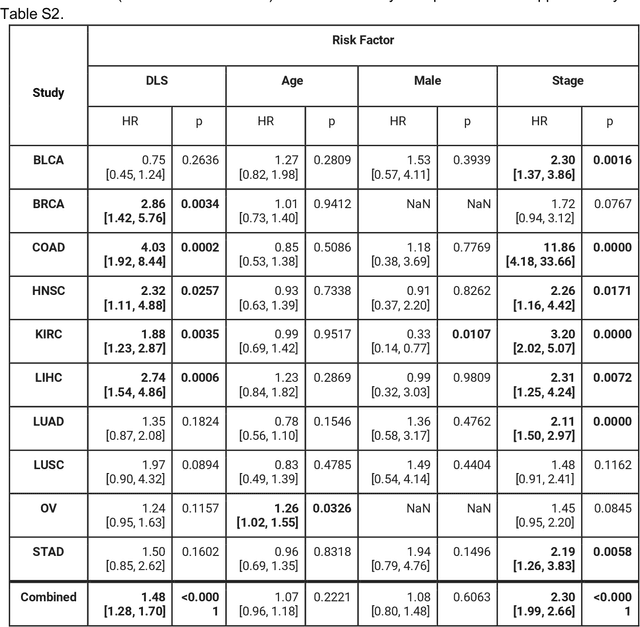
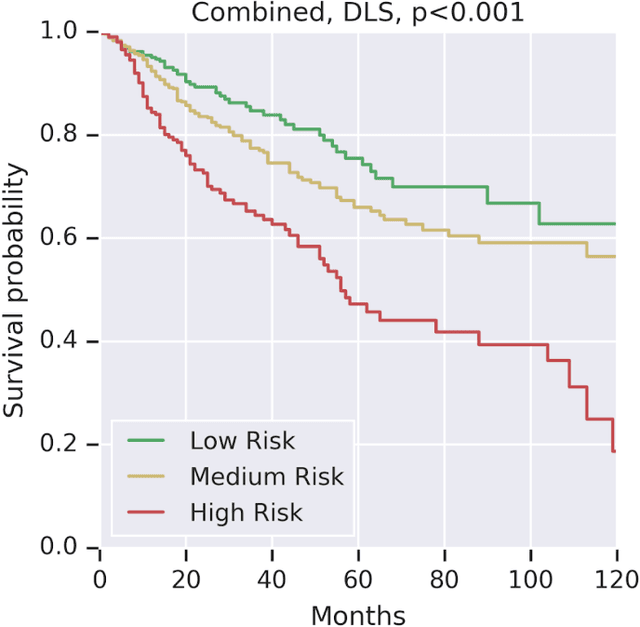
Abstract:Prognostic information at diagnosis has important implications for cancer treatment and monitoring. Although cancer staging, histopathological assessment, molecular features, and clinical variables can provide useful prognostic insights, improving risk stratification remains an active research area. We developed a deep learning system (DLS) to predict disease specific survival across 10 cancer types from The Cancer Genome Atlas (TCGA). We used a weakly-supervised approach without pixel-level annotations, and tested three different survival loss functions. The DLS was developed using 9,086 slides from 3,664 cases and evaluated using 3,009 slides from 1,216 cases. In multivariable Cox regression analysis of the combined cohort including all 10 cancers, the DLS was significantly associated with disease specific survival (hazard ratio of 1.58, 95% CI 1.28-1.70, p<0.0001) after adjusting for cancer type, stage, age, and sex. In a per-cancer adjusted subanalysis, the DLS remained a significant predictor of survival in 5 of 10 cancer types. Compared to a baseline model including stage, age, and sex, the c-index of the model demonstrated an absolute 3.7% improvement (95% CI 1.0-6.5) in the combined cohort. Additionally, our models stratified patients within individual cancer stages, particularly stage II (p=0.025) and stage III (p<0.001). By developing and evaluating prognostic models across multiple cancer types, this work represents one of the most comprehensive studies exploring the direct prediction of clinical outcomes using deep learning and histopathology images. Our analysis demonstrates the potential for this approach to provide prognostic information in multiple cancer types, and even within specific pathologic stages. However, given the relatively small number of clinical events, we observed wide confidence intervals, suggesting that future work will benefit from larger datasets.
Development and Validation of a Deep Learning Algorithm for Improving Gleason Scoring of Prostate Cancer
Nov 15, 2018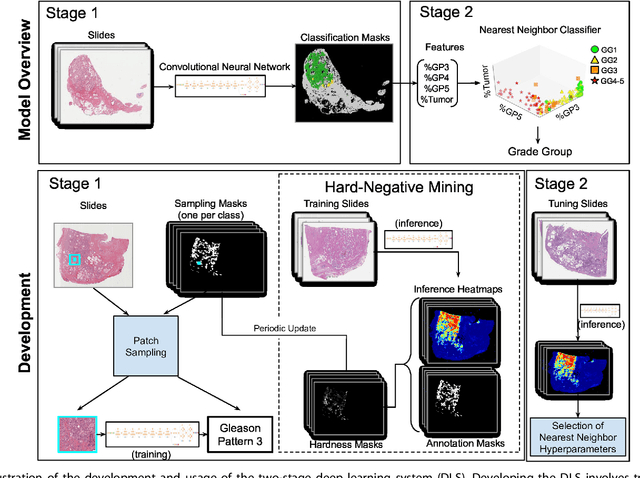
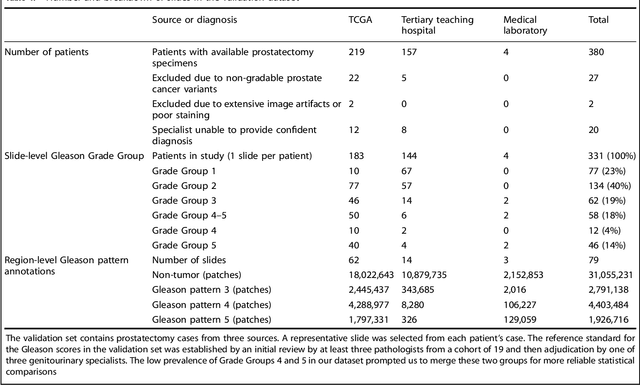
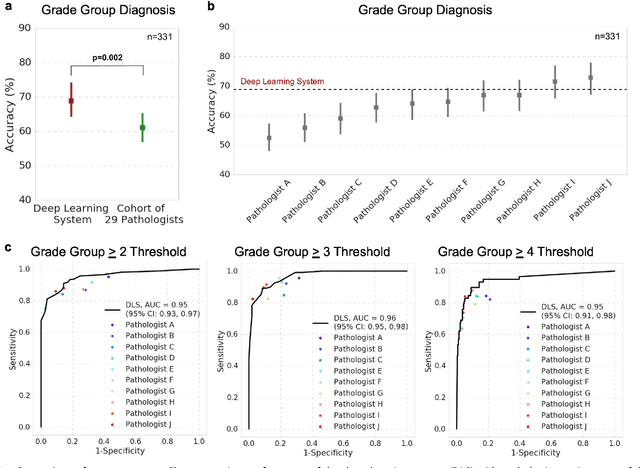
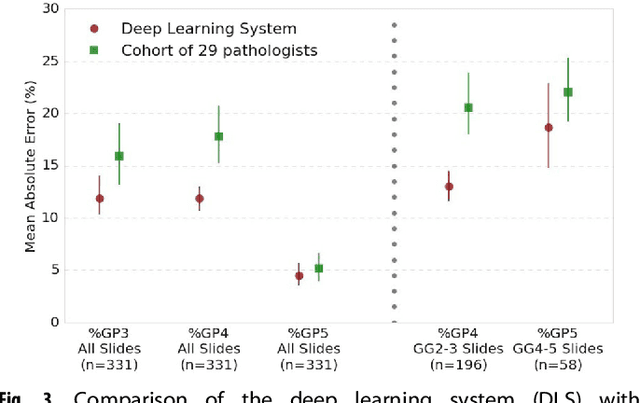
Abstract:For prostate cancer patients, the Gleason score is one of the most important prognostic factors, potentially determining treatment independent of the stage. However, Gleason scoring is based on subjective microscopic examination of tumor morphology and suffers from poor reproducibility. Here we present a deep learning system (DLS) for Gleason scoring whole-slide images of prostatectomies. Our system was developed using 112 million pathologist-annotated image patches from 1,226 slides, and evaluated on an independent validation dataset of 331 slides, where the reference standard was established by genitourinary specialist pathologists. On the validation dataset, the mean accuracy among 29 general pathologists was 0.61. The DLS achieved a significantly higher diagnostic accuracy of 0.70 (p=0.002) and trended towards better patient risk stratification in correlations to clinical follow-up data. Our approach could improve the accuracy of Gleason scoring and subsequent therapy decisions, particularly where specialist expertise is unavailable. The DLS also goes beyond the current Gleason system to more finely characterize and quantitate tumor morphology, providing opportunities for refinement of the Gleason system itself.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge