Yucheng Tang
VISTA-PATH: An interactive foundation model for pathology image segmentation and quantitative analysis in computational pathology
Jan 23, 2026Abstract:Accurate semantic segmentation for histopathology image is crucial for quantitative tissue analysis and downstream clinical modeling. Recent segmentation foundation models have improved generalization through large-scale pretraining, yet remain poorly aligned with pathology because they treat segmentation as a static visual prediction task. Here we present VISTA-PATH, an interactive, class-aware pathology segmentation foundation model designed to resolve heterogeneous structures, incorporate expert feedback, and produce pixel-level segmentation that are directly meaningful for clinical interpretation. VISTA-PATH jointly conditions segmentation on visual context, semantic tissue descriptions, and optional expert-provided spatial prompts, enabling precise multi-class segmentation across heterogeneous pathology images. To support this paradigm, we curate VISTA-PATH Data, a large-scale pathology segmentation corpus comprising over 1.6 million image-mask-text triplets spanning 9 organs and 93 tissue classes. Across extensive held-out and external benchmarks, VISTA-PATH consistently outperforms existing segmentation foundation models. Importantly, VISTA-PATH supports dynamic human-in-the-loop refinement by propagating sparse, patch-level bounding-box annotation feedback into whole-slide segmentation. Finally, we show that the high-fidelity, class-aware segmentation produced by VISTA-PATH is a preferred model for computational pathology. It improve tissue microenvironment analysis through proposed Tumor Interaction Score (TIS), which exhibits strong and significant associations with patient survival. Together, these results establish VISTA-PATH as a foundation model that elevates pathology image segmentation from a static prediction to an interactive and clinically grounded representation for digital pathology. Source code and demo can be found at https://github.com/zhihuanglab/VISTA-PATH.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
Img2ST-Net: Efficient High-Resolution Spatial Omics Prediction from Whole Slide Histology Images via Fully Convolutional Image-to-Image Learning
Aug 20, 2025



Abstract:Recent advances in multi-modal AI have demonstrated promising potential for generating the currently expensive spatial transcriptomics (ST) data directly from routine histology images, offering a means to reduce the high cost and time-intensive nature of ST data acquisition. However, the increasing resolution of ST, particularly with platforms such as Visium HD achieving 8um or finer, introduces significant computational and modeling challenges. Conventional spot-by-spot sequential regression frameworks become inefficient and unstable at this scale, while the inherent extreme sparsity and low expression levels of high-resolution ST further complicate both prediction and evaluation. To address these limitations, we propose Img2ST-Net, a novel histology-to-ST generation framework for efficient and parallel high-resolution ST prediction. Unlike conventional spot-by-spot inference methods, Img2ST-Net employs a fully convolutional architecture to generate dense, HD gene expression maps in a parallelized manner. By modeling HD ST data as super-pixel representations, the task is reformulated from image-to-omics inference into a super-content image generation problem with hundreds or thousands of output channels. This design not only improves computational efficiency but also better preserves the spatial organization intrinsic to spatial omics data. To enhance robustness under sparse expression patterns, we further introduce SSIM-ST, a structural-similarity-based evaluation metric tailored for high-resolution ST analysis. We present a scalable, biologically coherent framework for high-resolution ST prediction. Img2ST-Net offers a principled solution for efficient and accurate ST inference at scale. Our contributions lay the groundwork for next-generation ST modeling that is robust and resolution-aware. The source code has been made publicly available at https://github.com/hrlblab/Img2ST-Net.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
BEAST: Efficient Tokenization of B-Splines Encoded Action Sequences for Imitation Learning
Jun 06, 2025Abstract:We present the B-spline Encoded Action Sequence Tokenizer (BEAST), a novel action tokenizer that encodes action sequences into compact discrete or continuous tokens using B-splines. In contrast to existing action tokenizers based on vector quantization or byte pair encoding, BEAST requires no separate tokenizer training and consistently produces tokens of uniform length, enabling fast action sequence generation via parallel decoding. Leveraging our B-spline formulation, BEAST inherently ensures generating smooth trajectories without discontinuities between adjacent segments. We extensively evaluate BEAST by integrating it with three distinct model architectures: a Variational Autoencoder (VAE) with continuous tokens, a decoder-only Transformer with discrete tokens, and Florence-2, a pretrained Vision-Language Model with an encoder-decoder architecture, demonstrating BEAST's compatibility and scalability with large pretrained models. We evaluate BEAST across three established benchmarks consisting of 166 simulated tasks and on three distinct robot settings with a total of 8 real-world tasks. Experimental results demonstrate that BEAST (i) significantly reduces both training and inference computational costs, and (ii) consistently generates smooth, high-frequency control signals suitable for continuous control tasks while (iii) reliably achieves competitive task success rates compared to state-of-the-art methods.
IRS: Incremental Relationship-guided Segmentation for Digital Pathology
May 28, 2025Abstract:Continual learning is rapidly emerging as a key focus in computer vision, aiming to develop AI systems capable of continuous improvement, thereby enhancing their value and practicality in diverse real-world applications. In healthcare, continual learning holds great promise for continuously acquired digital pathology data, which is collected in hospitals on a daily basis. However, panoramic segmentation on digital whole slide images (WSIs) presents significant challenges, as it is often infeasible to obtain comprehensive annotations for all potential objects, spanning from coarse structures (e.g., regions and unit objects) to fine structures (e.g., cells). This results in temporally and partially annotated data, posing a major challenge in developing a holistic segmentation framework. Moreover, an ideal segmentation model should incorporate new phenotypes, unseen diseases, and diverse populations, making this task even more complex. In this paper, we introduce a novel and unified Incremental Relationship-guided Segmentation (IRS) learning scheme to address temporally acquired, partially annotated data while maintaining out-of-distribution (OOD) continual learning capacity in digital pathology. The key innovation of IRS lies in its ability to realize a new spatial-temporal OOD continual learning paradigm by mathematically modeling anatomical relationships between existing and newly introduced classes through a simple incremental universal proposition matrix. Experimental results demonstrate that the IRS method effectively handles the multi-scale nature of pathological segmentation, enabling precise kidney segmentation across various structures (regions, units, and cells) as well as OOD disease lesions at multiple magnifications. This capability significantly enhances domain generalization, making IRS a robust approach for real-world digital pathology applications.
MedSegFactory: Text-Guided Generation of Medical Image-Mask Pairs
Apr 09, 2025Abstract:This paper presents MedSegFactory, a versatile medical synthesis framework that generates high-quality paired medical images and segmentation masks across modalities and tasks. It aims to serve as an unlimited data repository, supplying image-mask pairs to enhance existing segmentation tools. The core of MedSegFactory is a dual-stream diffusion model, where one stream synthesizes medical images and the other generates corresponding segmentation masks. To ensure precise alignment between image-mask pairs, we introduce Joint Cross-Attention (JCA), enabling a collaborative denoising paradigm by dynamic cross-conditioning between streams. This bidirectional interaction allows both representations to guide each other's generation, enhancing consistency between generated pairs. MedSegFactory unlocks on-demand generation of paired medical images and segmentation masks through user-defined prompts that specify the target labels, imaging modalities, anatomical regions, and pathological conditions, facilitating scalable and high-quality data generation. This new paradigm of medical image synthesis enables seamless integration into diverse medical imaging workflows, enhancing both efficiency and accuracy. Extensive experiments show that MedSegFactory generates data of superior quality and usability, achieving competitive or state-of-the-art performance in 2D and 3D segmentation tasks while addressing data scarcity and regulatory constraints.
Interactive Tumor Progression Modeling via Sketch-Based Image Editing
Mar 10, 2025



Abstract:Accurately visualizing and editing tumor progression in medical imaging is crucial for diagnosis, treatment planning, and clinical communication. To address the challenges of subjectivity and limited precision in existing methods, we propose SkEditTumor, a sketch-based diffusion model for controllable tumor progression editing. By leveraging sketches as structural priors, our method enables precise modifications of tumor regions while maintaining structural integrity and visual realism. We evaluate SkEditTumor on four public datasets - BraTS, LiTS, KiTS, and MSD-Pancreas - covering diverse organs and imaging modalities. Experimental results demonstrate that our method outperforms state-of-the-art baselines, achieving superior image fidelity and segmentation accuracy. Our contributions include a novel integration of sketches with diffusion models for medical image editing, fine-grained control over tumor progression visualization, and extensive validation across multiple datasets, setting a new benchmark in the field.
MagNet: Multi-Level Attention Graph Network for Predicting High-Resolution Spatial Transcriptomics
Feb 28, 2025



Abstract:The rapid development of spatial transcriptomics (ST) offers new opportunities to explore the gene expression patterns within the spatial microenvironment. Current research integrates pathological images to infer gene expression, addressing the high costs and time-consuming processes to generate spatial transcriptomics data. However, as spatial transcriptomics resolution continues to improve, existing methods remain primarily focused on gene expression prediction at low-resolution spot levels. These methods face significant challenges, especially the information bottleneck, when they are applied to high-resolution HD data. To bridge this gap, this paper introduces MagNet, a multi-level attention graph network designed for accurate prediction of high-resolution HD data. MagNet employs cross-attention layers to integrate features from multi-resolution image patches hierarchically and utilizes a GAT-Transformer module to aggregate neighborhood information. By integrating multilevel features, MagNet overcomes the limitations posed by low-resolution inputs in predicting high-resolution gene expression. We systematically evaluated MagNet and existing ST prediction models on both a private spatial transcriptomics dataset and a public dataset at three different resolution levels. The results demonstrate that MagNet achieves state-of-the-art performance at both spot level and high-resolution bin levels, providing a novel methodology and benchmark for future research and applications in high-resolution HD-level spatial transcriptomics. Code is available at https://github.com/Junchao-Zhu/MagNet.
KPIs 2024 Challenge: Advancing Glomerular Segmentation from Patch- to Slide-Level
Feb 11, 2025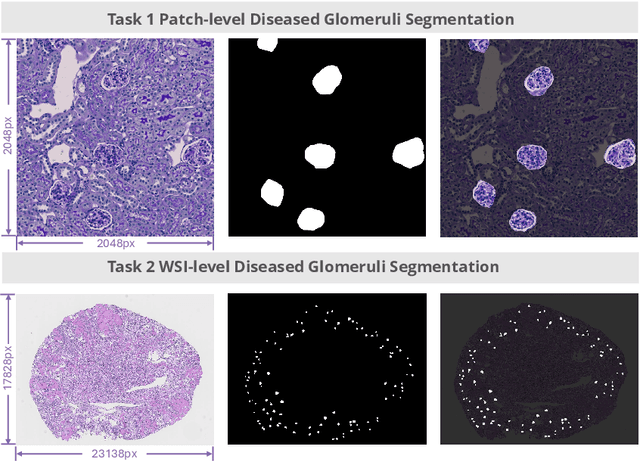
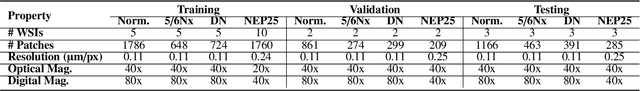
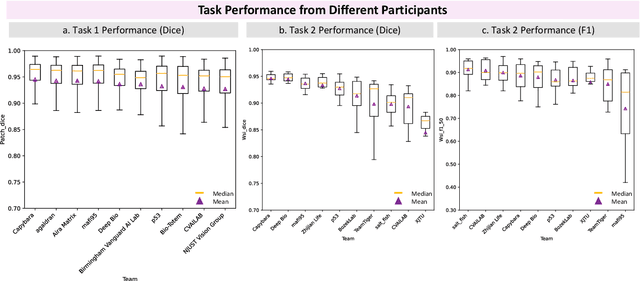

Abstract:Chronic kidney disease (CKD) is a major global health issue, affecting over 10% of the population and causing significant mortality. While kidney biopsy remains the gold standard for CKD diagnosis and treatment, the lack of comprehensive benchmarks for kidney pathology segmentation hinders progress in the field. To address this, we organized the Kidney Pathology Image Segmentation (KPIs) Challenge, introducing a dataset that incorporates preclinical rodent models of CKD with over 10,000 annotated glomeruli from 60+ Periodic Acid Schiff (PAS)-stained whole slide images. The challenge includes two tasks, patch-level segmentation and whole slide image segmentation and detection, evaluated using the Dice Similarity Coefficient (DSC) and F1-score. By encouraging innovative segmentation methods that adapt to diverse CKD models and tissue conditions, the KPIs Challenge aims to advance kidney pathology analysis, establish new benchmarks, and enable precise, large-scale quantification for disease research and diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge