Siqi Lu
SCR2-ST: Combine Single Cell with Spatial Transcriptomics for Efficient Active Sampling via Reinforcement Learning
Dec 15, 2025



Abstract:Spatial transcriptomics (ST) is an emerging technology that enables researchers to investigate the molecular relationships underlying tissue morphology. However, acquiring ST data remains prohibitively expensive, and traditional fixed-grid sampling strategies lead to redundant measurements of morphologically similar or biologically uninformative regions, thus resulting in scarce data that constrain current methods. The well-established single-cell sequencing field, however, could provide rich biological data as an effective auxiliary source to mitigate this limitation. To bridge these gaps, we introduce SCR2-ST, a unified framework that leverages single-cell prior knowledge to guide efficient data acquisition and accurate expression prediction. SCR2-ST integrates a single-cell guided reinforcement learning-based (SCRL) active sampling and a hybrid regression-retrieval prediction network SCR2Net. SCRL combines single-cell foundation model embeddings with spatial density information to construct biologically grounded reward signals, enabling selective acquisition of informative tissue regions under constrained sequencing budgets. SCR2Net then leverages the actively sampled data through a hybrid architecture combining regression-based modeling with retrieval-augmented inference, where a majority cell-type filtering mechanism suppresses noisy matches and retrieved expression profiles serve as soft labels for auxiliary supervision. We evaluated SCR2-ST on three public ST datasets, demonstrating SOTA performance in both sampling efficiency and prediction accuracy, particularly under low-budget scenarios. Code is publicly available at: https://github.com/hrlblab/SCR2ST
IRS: Incremental Relationship-guided Segmentation for Digital Pathology
May 28, 2025Abstract:Continual learning is rapidly emerging as a key focus in computer vision, aiming to develop AI systems capable of continuous improvement, thereby enhancing their value and practicality in diverse real-world applications. In healthcare, continual learning holds great promise for continuously acquired digital pathology data, which is collected in hospitals on a daily basis. However, panoramic segmentation on digital whole slide images (WSIs) presents significant challenges, as it is often infeasible to obtain comprehensive annotations for all potential objects, spanning from coarse structures (e.g., regions and unit objects) to fine structures (e.g., cells). This results in temporally and partially annotated data, posing a major challenge in developing a holistic segmentation framework. Moreover, an ideal segmentation model should incorporate new phenotypes, unseen diseases, and diverse populations, making this task even more complex. In this paper, we introduce a novel and unified Incremental Relationship-guided Segmentation (IRS) learning scheme to address temporally acquired, partially annotated data while maintaining out-of-distribution (OOD) continual learning capacity in digital pathology. The key innovation of IRS lies in its ability to realize a new spatial-temporal OOD continual learning paradigm by mathematically modeling anatomical relationships between existing and newly introduced classes through a simple incremental universal proposition matrix. Experimental results demonstrate that the IRS method effectively handles the multi-scale nature of pathological segmentation, enabling precise kidney segmentation across various structures (regions, units, and cells) as well as OOD disease lesions at multiple magnifications. This capability significantly enhances domain generalization, making IRS a robust approach for real-world digital pathology applications.
DeepAndes: A Self-Supervised Vision Foundation Model for Multi-Spectral Remote Sensing Imagery of the Andes
Apr 28, 2025Abstract:By mapping sites at large scales using remotely sensed data, archaeologists can generate unique insights into long-term demographic trends, inter-regional social networks, and past adaptations to climate change. Remote sensing surveys complement field-based approaches, and their reach can be especially great when combined with deep learning and computer vision techniques. However, conventional supervised deep learning methods face challenges in annotating fine-grained archaeological features at scale. While recent vision foundation models have shown remarkable success in learning large-scale remote sensing data with minimal annotations, most off-the-shelf solutions are designed for RGB images rather than multi-spectral satellite imagery, such as the 8-band data used in our study. In this paper, we introduce DeepAndes, a transformer-based vision foundation model trained on three million multi-spectral satellite images, specifically tailored for Andean archaeology. DeepAndes incorporates a customized DINOv2 self-supervised learning algorithm optimized for 8-band multi-spectral imagery, marking the first foundation model designed explicitly for the Andes region. We evaluate its image understanding performance through imbalanced image classification, image instance retrieval, and pixel-level semantic segmentation tasks. Our experiments show that DeepAndes achieves superior F1 scores, mean average precision, and Dice scores in few-shot learning scenarios, significantly outperforming models trained from scratch or pre-trained on smaller datasets. This underscores the effectiveness of large-scale self-supervised pre-training in archaeological remote sensing. Codes will be available on https://github.com/geopacha/DeepAndes.
MagNet: Multi-Level Attention Graph Network for Predicting High-Resolution Spatial Transcriptomics
Feb 28, 2025



Abstract:The rapid development of spatial transcriptomics (ST) offers new opportunities to explore the gene expression patterns within the spatial microenvironment. Current research integrates pathological images to infer gene expression, addressing the high costs and time-consuming processes to generate spatial transcriptomics data. However, as spatial transcriptomics resolution continues to improve, existing methods remain primarily focused on gene expression prediction at low-resolution spot levels. These methods face significant challenges, especially the information bottleneck, when they are applied to high-resolution HD data. To bridge this gap, this paper introduces MagNet, a multi-level attention graph network designed for accurate prediction of high-resolution HD data. MagNet employs cross-attention layers to integrate features from multi-resolution image patches hierarchically and utilizes a GAT-Transformer module to aggregate neighborhood information. By integrating multilevel features, MagNet overcomes the limitations posed by low-resolution inputs in predicting high-resolution gene expression. We systematically evaluated MagNet and existing ST prediction models on both a private spatial transcriptomics dataset and a public dataset at three different resolution levels. The results demonstrate that MagNet achieves state-of-the-art performance at both spot level and high-resolution bin levels, providing a novel methodology and benchmark for future research and applications in high-resolution HD-level spatial transcriptomics. Code is available at https://github.com/Junchao-Zhu/MagNet.
KPIs 2024 Challenge: Advancing Glomerular Segmentation from Patch- to Slide-Level
Feb 11, 2025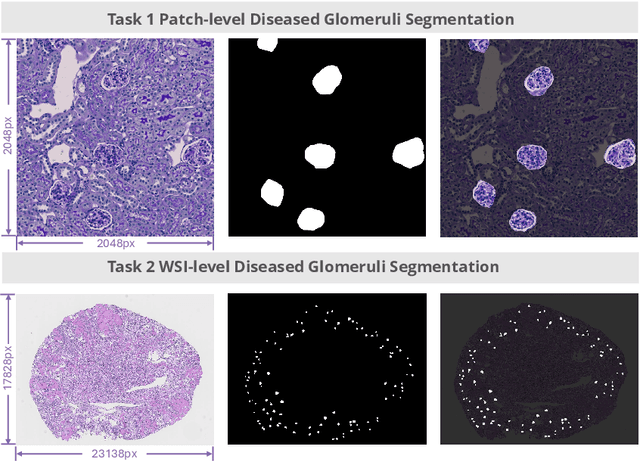
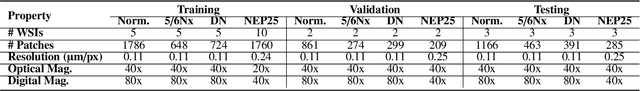
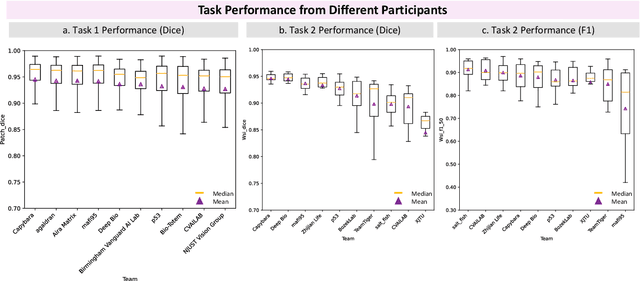

Abstract:Chronic kidney disease (CKD) is a major global health issue, affecting over 10% of the population and causing significant mortality. While kidney biopsy remains the gold standard for CKD diagnosis and treatment, the lack of comprehensive benchmarks for kidney pathology segmentation hinders progress in the field. To address this, we organized the Kidney Pathology Image Segmentation (KPIs) Challenge, introducing a dataset that incorporates preclinical rodent models of CKD with over 10,000 annotated glomeruli from 60+ Periodic Acid Schiff (PAS)-stained whole slide images. The challenge includes two tasks, patch-level segmentation and whole slide image segmentation and detection, evaluated using the Dice Similarity Coefficient (DSC) and F1-score. By encouraging innovative segmentation methods that adapt to diverse CKD models and tissue conditions, the KPIs Challenge aims to advance kidney pathology analysis, establish new benchmarks, and enable precise, large-scale quantification for disease research and diagnosis.
ASIGN: An Anatomy-aware Spatial Imputation Graphic Network for 3D Spatial Transcriptomics
Dec 04, 2024



Abstract:Spatial transcriptomics (ST) is an emerging technology that enables medical computer vision scientists to automatically interpret the molecular profiles underlying morphological features. Currently, however, most deep learning-based ST analyses are limited to two-dimensional (2D) sections, which can introduce diagnostic errors due to the heterogeneity of pathological tissues across 3D sections. Expanding ST to three-dimensional (3D) volumes is challenging due to the prohibitive costs; a 2D ST acquisition already costs over 50 times more than whole slide imaging (WSI), and a full 3D volume with 10 sections can be an order of magnitude more expensive. To reduce costs, scientists have attempted to predict ST data directly from WSI without performing actual ST acquisition. However, these methods typically yield unsatisfying results. To address this, we introduce a novel problem setting: 3D ST imputation using 3D WSI histology sections combined with a single 2D ST slide. To do so, we present the Anatomy-aware Spatial Imputation Graph Network (ASIGN) for more precise, yet affordable, 3D ST modeling. The ASIGN architecture extends existing 2D spatial relationships into 3D by leveraging cross-layer overlap and similarity-based expansion. Moreover, a multi-level spatial attention graph network integrates features comprehensively across different data sources. We evaluated ASIGN on three public spatial transcriptomics datasets, with experimental results demonstrating that ASIGN achieves state-of-the-art performance on both 2D and 3D scenarios. Code is available at https://github.com/hrlblab/ASIGN.
GloFinder: AI-empowered QuPath Plugin for WSI-level Glomerular Detection, Visualization, and Curation
Nov 27, 2024


Abstract:Artificial intelligence (AI) has demonstrated significant success in automating the detection of glomeruli, the key functional units of the kidney, from whole slide images (WSIs) in kidney pathology. However, existing open-source tools are often distributed as source code or Docker containers, requiring advanced programming skills that hinder accessibility for non-programmers, such as clinicians. Additionally, current models are typically trained on a single dataset and lack flexibility in adjusting confidence levels for predictions. To overcome these challenges, we introduce GloFinder, a QuPath plugin designed for single-click automated glomeruli detection across entire WSIs with online editing through the graphical user interface (GUI). GloFinder employs CircleNet, an anchor-free detection framework utilizing circle representations for precise object localization, with models trained on approximately 160,000 manually annotated glomeruli. To further enhance accuracy, the plugin incorporates Weighted Circle Fusion (WCF), an ensemble method that combines confidence scores from multiple CircleNet models to produce refined predictions, achieving superior performance in glomerular detection. GloFinder enables direct visualization and editing of results in QuPath, facilitating seamless interaction for clinicians and providing a powerful tool for nephropathology research and clinical practice.
How Good Are We? Evaluating Cell AI Foundation Models in Kidney Pathology with Human-in-the-Loop Enrichment
Oct 31, 2024



Abstract:Training AI foundation models has emerged as a promising large-scale learning approach for addressing real-world healthcare challenges, including digital pathology. While many of these models have been developed for tasks like disease diagnosis and tissue quantification using extensive and diverse training datasets, their readiness for deployment on some arguably simplest tasks, such as nuclei segmentation within a single organ (e.g., the kidney), remains uncertain. This paper seeks to answer this key question, "How good are we?", by thoroughly evaluating the performance of recent cell foundation models on a curated multi-center, multi-disease, and multi-species external testing dataset. Additionally, we tackle a more challenging question, "How can we improve?", by developing and assessing human-in-the-loop data enrichment strategies aimed at enhancing model performance while minimizing the reliance on pixel-level human annotation. To address the first question, we curated a multicenter, multidisease, and multispecies dataset consisting of 2,542 kidney whole slide images (WSIs). Three state-of-the-art (SOTA) cell foundation models-Cellpose, StarDist, and CellViT-were selected for evaluation. To tackle the second question, we explored data enrichment algorithms by distilling predictions from the different foundation models with a human-in-the-loop framework, aiming to further enhance foundation model performance with minimal human efforts. Our experimental results showed that all three foundation models improved over their baselines with model fine-tuning with enriched data. Interestingly, the baseline model with the highest F1 score does not yield the best segmentation outcomes after fine-tuning. This study establishes a benchmark for the development and deployment of cell vision foundation models tailored for real-world data applications.
Assessment of Cell Nuclei AI Foundation Models in Kidney Pathology
Aug 09, 2024Abstract:Cell nuclei instance segmentation is a crucial task in digital kidney pathology. Traditional automatic segmentation methods often lack generalizability when applied to unseen datasets. Recently, the success of foundation models (FMs) has provided a more generalizable solution, potentially enabling the segmentation of any cell type. In this study, we perform a large-scale evaluation of three widely used state-of-the-art (SOTA) cell nuclei foundation models (Cellpose, StarDist, and CellViT). Specifically, we created a highly diverse evaluation dataset consisting of 2,542 kidney whole slide images (WSIs) collected from both human and rodent sources, encompassing various tissue types, sizes, and staining methods. To our knowledge, this is the largest-scale evaluation of its kind to date. Our quantitative analysis of the prediction distribution reveals a persistent performance gap in kidney pathology. Among the evaluated models, CellViT demonstrated superior performance in segmenting nuclei in kidney pathology. However, none of the foundation models are perfect; a performance gap remains in general nuclei segmentation for kidney pathology.
AI Foundation Models in Remote Sensing: A Survey
Aug 06, 2024Abstract:Artificial Intelligence (AI) technologies have profoundly transformed the field of remote sensing, revolutionizing data collection, processing, and analysis. Traditionally reliant on manual interpretation and task-specific models, remote sensing has been significantly enhanced by the advent of foundation models--large-scale, pre-trained AI models capable of performing a wide array of tasks with unprecedented accuracy and efficiency. This paper provides a comprehensive survey of foundation models in the remote sensing domain, covering models released between June 2021 and June 2024. We categorize these models based on their applications in computer vision and domain-specific tasks, offering insights into their architectures, pre-training datasets, and methodologies. Through detailed performance comparisons, we highlight emerging trends and the significant advancements achieved by these foundation models. Additionally, we discuss the technical challenges, practical implications, and future research directions, addressing the need for high-quality data, computational resources, and improved model generalization. Our research also finds that pre-training methods, particularly self-supervised learning techniques like contrastive learning and masked autoencoders, significantly enhance the performance and robustness of foundation models in remote sensing tasks such as scene classification, object detection, and other applications. This survey aims to serve as a resource for researchers and practitioners by providing a panorama of advances and promising pathways for continued development and application of foundation models in remote sensing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge