Xiaoxi Chen
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
Baichuan-Omni-1.5 Technical Report
Jan 26, 2025
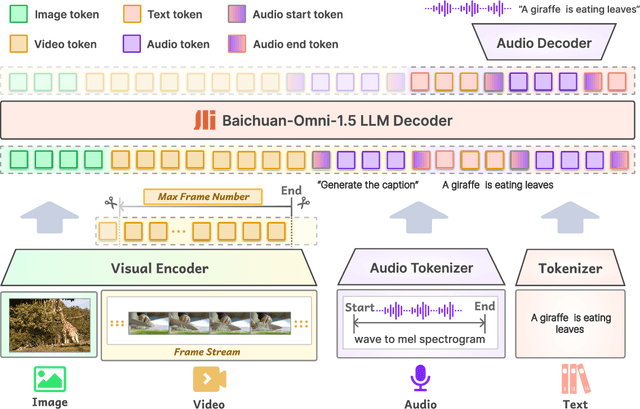
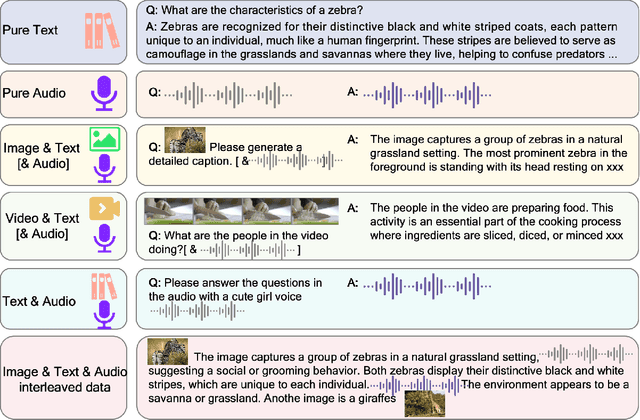
Abstract:We introduce Baichuan-Omni-1.5, an omni-modal model that not only has omni-modal understanding capabilities but also provides end-to-end audio generation capabilities. To achieve fluent and high-quality interaction across modalities without compromising the capabilities of any modality, we prioritized optimizing three key aspects. First, we establish a comprehensive data cleaning and synthesis pipeline for multimodal data, obtaining about 500B high-quality data (text, audio, and vision). Second, an audio-tokenizer (Baichuan-Audio-Tokenizer) has been designed to capture both semantic and acoustic information from audio, enabling seamless integration and enhanced compatibility with MLLM. Lastly, we designed a multi-stage training strategy that progressively integrates multimodal alignment and multitask fine-tuning, ensuring effective synergy across all modalities. Baichuan-Omni-1.5 leads contemporary models (including GPT4o-mini and MiniCPM-o 2.6) in terms of comprehensive omni-modal capabilities. Notably, it achieves results comparable to leading models such as Qwen2-VL-72B across various multimodal medical benchmarks.
RadGPT: Constructing 3D Image-Text Tumor Datasets
Jan 08, 2025Abstract:With over 85 million CT scans performed annually in the United States, creating tumor-related reports is a challenging and time-consuming task for radiologists. To address this need, we present RadGPT, an Anatomy-Aware Vision-Language AI Agent for generating detailed reports from CT scans. RadGPT first segments tumors, including benign cysts and malignant tumors, and their surrounding anatomical structures, then transforms this information into both structured reports and narrative reports. These reports provide tumor size, shape, location, attenuation, volume, and interactions with surrounding blood vessels and organs. Extensive evaluation on unseen hospitals shows that RadGPT can produce accurate reports, with high sensitivity/specificity for small tumor (<2 cm) detection: 80/73% for liver tumors, 92/78% for kidney tumors, and 77/77% for pancreatic tumors. For large tumors, sensitivity ranges from 89% to 97%. The results significantly surpass the state-of-the-art in abdominal CT report generation. RadGPT generated reports for 17 public datasets. Through radiologist review and refinement, we have ensured the reports' accuracy, and created the first publicly available image-text 3D medical dataset, comprising over 1.8 million text tokens and 2.7 million images from 9,262 CT scans, including 2,947 tumor scans/reports of 8,562 tumor instances. Our reports can: (1) localize tumors in eight liver sub-segments and three pancreatic sub-segments annotated per-voxel; (2) determine pancreatic tumor stage (T1-T4) in 260 reports; and (3) present individual analyses of multiple tumors--rare in human-made reports. Importantly, 948 of the reports are for early-stage tumors.
ScaleMAI: Accelerating the Development of Trusted Datasets and AI Models
Jan 06, 2025



Abstract:Building trusted datasets is critical for transparent and responsible Medical AI (MAI) research, but creating even small, high-quality datasets can take years of effort from multidisciplinary teams. This process often delays AI benefits, as human-centric data creation and AI-centric model development are treated as separate, sequential steps. To overcome this, we propose ScaleMAI, an agent of AI-integrated data curation and annotation, allowing data quality and AI performance to improve in a self-reinforcing cycle and reducing development time from years to months. We adopt pancreatic tumor detection as an example. First, ScaleMAI progressively creates a dataset of 25,362 CT scans, including per-voxel annotations for benign/malignant tumors and 24 anatomical structures. Second, through progressive human-in-the-loop iterations, ScaleMAI provides Flagship AI Model that can approach the proficiency of expert annotators (30-year experience) in detecting pancreatic tumors. Flagship Model significantly outperforms models developed from smaller, fixed-quality datasets, with substantial gains in tumor detection (+14%), segmentation (+5%), and classification (72%) on three prestigious benchmarks. In summary, ScaleMAI transforms the speed, scale, and reliability of medical dataset creation, paving the way for a variety of impactful, data-driven applications.
Analyzing Tumors by Synthesis
Sep 09, 2024



Abstract:Computer-aided tumor detection has shown great potential in enhancing the interpretation of over 80 million CT scans performed annually in the United States. However, challenges arise due to the rarity of CT scans with tumors, especially early-stage tumors. Developing AI with real tumor data faces issues of scarcity, annotation difficulty, and low prevalence. Tumor synthesis addresses these challenges by generating numerous tumor examples in medical images, aiding AI training for tumor detection and segmentation. Successful synthesis requires realistic and generalizable synthetic tumors across various organs. This chapter reviews AI development on real and synthetic data and summarizes two key trends in synthetic data for cancer imaging research: modeling-based and learning-based approaches. Modeling-based methods, like Pixel2Cancer, simulate tumor development over time using generic rules, while learning-based methods, like DiffTumor, learn from a few annotated examples in one organ to generate synthetic tumors in others. Reader studies with expert radiologists show that synthetic tumors can be convincingly realistic. We also present case studies in the liver, pancreas, and kidneys reveal that AI trained on synthetic tumors can achieve performance comparable to, or better than, AI only trained on real data. Tumor synthesis holds significant promise for expanding datasets, enhancing AI reliability, improving tumor detection performance, and preserving patient privacy.
AbdomenAtlas: A Large-Scale, Detailed-Annotated, & Multi-Center Dataset for Efficient Transfer Learning and Open Algorithmic Benchmarking
Jul 23, 2024



Abstract:We introduce the largest abdominal CT dataset (termed AbdomenAtlas) of 20,460 three-dimensional CT volumes sourced from 112 hospitals across diverse populations, geographies, and facilities. AbdomenAtlas provides 673K high-quality masks of anatomical structures in the abdominal region annotated by a team of 10 radiologists with the help of AI algorithms. We start by having expert radiologists manually annotate 22 anatomical structures in 5,246 CT volumes. Following this, a semi-automatic annotation procedure is performed for the remaining CT volumes, where radiologists revise the annotations predicted by AI, and in turn, AI improves its predictions by learning from revised annotations. Such a large-scale, detailed-annotated, and multi-center dataset is needed for two reasons. Firstly, AbdomenAtlas provides important resources for AI development at scale, branded as large pre-trained models, which can alleviate the annotation workload of expert radiologists to transfer to broader clinical applications. Secondly, AbdomenAtlas establishes a large-scale benchmark for evaluating AI algorithms -- the more data we use to test the algorithms, the better we can guarantee reliable performance in complex clinical scenarios. An ISBI & MICCAI challenge named BodyMaps: Towards 3D Atlas of Human Body was launched using a subset of our AbdomenAtlas, aiming to stimulate AI innovation and to benchmark segmentation accuracy, inference efficiency, and domain generalizability. We hope our AbdomenAtlas can set the stage for larger-scale clinical trials and offer exceptional opportunities to practitioners in the medical imaging community. Codes, models, and datasets are available at https://www.zongweiz.com/dataset
Universal and Extensible Language-Vision Models for Organ Segmentation and Tumor Detection from Abdominal Computed Tomography
May 28, 2024



Abstract:The advancement of artificial intelligence (AI) for organ segmentation and tumor detection is propelled by the growing availability of computed tomography (CT) datasets with detailed, per-voxel annotations. However, these AI models often struggle with flexibility for partially annotated datasets and extensibility for new classes due to limitations in the one-hot encoding, architectural design, and learning scheme. To overcome these limitations, we propose a universal, extensible framework enabling a single model, termed Universal Model, to deal with multiple public datasets and adapt to new classes (e.g., organs/tumors). Firstly, we introduce a novel language-driven parameter generator that leverages language embeddings from large language models, enriching semantic encoding compared with one-hot encoding. Secondly, the conventional output layers are replaced with lightweight, class-specific heads, allowing Universal Model to simultaneously segment 25 organs and six types of tumors and ease the addition of new classes. We train our Universal Model on 3,410 CT volumes assembled from 14 publicly available datasets and then test it on 6,173 CT volumes from four external datasets. Universal Model achieves first place on six CT tasks in the Medical Segmentation Decathlon (MSD) public leaderboard and leading performance on the Beyond The Cranial Vault (BTCV) dataset. In summary, Universal Model exhibits remarkable computational efficiency (6x faster than other dataset-specific models), demonstrates strong generalization across different hospitals, transfers well to numerous downstream tasks, and more importantly, facilitates the extensibility to new classes while alleviating the catastrophic forgetting of previously learned classes. Codes, models, and datasets are available at https://github.com/ljwztc/CLIP-Driven-Universal-Model
From Pixel to Cancer: Cellular Automata in Computed Tomography
Mar 11, 2024Abstract:AI for cancer detection encounters the bottleneck of data scarcity, annotation difficulty, and low prevalence of early tumors. Tumor synthesis seeks to create artificial tumors in medical images, which can greatly diversify the data and annotations for AI training. However, current tumor synthesis approaches are not applicable across different organs due to their need for specific expertise and design. This paper establishes a set of generic rules to simulate tumor development. Each cell (pixel) is initially assigned a state between zero and ten to represent the tumor population, and a tumor can be developed based on three rules to describe the process of growth, invasion, and death. We apply these three generic rules to simulate tumor development--from pixel to cancer--using cellular automata. We then integrate the tumor state into the original computed tomography (CT) images to generate synthetic tumors across different organs. This tumor synthesis approach allows for sampling tumors at multiple stages and analyzing tumor-organ interaction. Clinically, a reader study involving three expert radiologists reveals that the synthetic tumors and their developing trajectories are convincingly realistic. Technically, we generate tumors at varied stages in 9,262 raw, unlabeled CT images sourced from 68 hospitals worldwide. The performance in segmenting tumors in the liver, pancreas, and kidneys exceeds prevailing literature benchmarks, underlining the immense potential of tumor synthesis, especially for earlier cancer detection. The code and models are available at https://github.com/MrGiovanni/Pixel2Cancer
Towards Generalizable Tumor Synthesis
Feb 29, 2024Abstract:Tumor synthesis enables the creation of artificial tumors in medical images, facilitating the training of AI models for tumor detection and segmentation. However, success in tumor synthesis hinges on creating visually realistic tumors that are generalizable across multiple organs and, furthermore, the resulting AI models being capable of detecting real tumors in images sourced from different domains (e.g., hospitals). This paper made a progressive stride toward generalizable tumor synthesis by leveraging a critical observation: early-stage tumors (< 2cm) tend to have similar imaging characteristics in computed tomography (CT), whether they originate in the liver, pancreas, or kidneys. We have ascertained that generative AI models, e.g., Diffusion Models, can create realistic tumors generalized to a range of organs even when trained on a limited number of tumor examples from only one organ. Moreover, we have shown that AI models trained on these synthetic tumors can be generalized to detect and segment real tumors from CT volumes, encompassing a broad spectrum of patient demographics, imaging protocols, and healthcare facilities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge