Xiyue Wang
From Tags to Trees: Structuring Fine-Grained Knowledge for Controllable Data Selection in LLM Instruction Tuning
Jan 20, 2026Abstract:Effective and controllable data selection is critical for LLM instruction tuning, especially with massive open-source datasets. Existing approaches primarily rely on instance-level quality scores, or diversity metrics based on embedding clusters or semantic tags. However, constrained by the flatness of embedding spaces or the coarseness of tags, these approaches overlook fine-grained knowledge and its intrinsic hierarchical dependencies, consequently hindering precise data valuation and knowledge-aligned sampling. To address this challenge, we propose Tree-aware Aligned Global Sampling (TAGS), a unified framework that leverages a knowledge tree built from fine-grained tags, thereby enabling joint control of global quality, diversity, and target alignment. Using an LLM-based tagger, we extract atomic knowledge concepts, which are organized into a global tree through bottom-up hierarchical clustering. By grounding data instances onto this tree, a tree-aware metric then quantifies data quality and diversity, facilitating effective sampling. Our controllable sampling strategy maximizes tree-level information gain and enforces leaf-level alignment via KL-divergence for specific domains. Extensive experiments demonstrate that TAGS significantly outperforms state-of-the-art baselines. Notably, it surpasses the full-dataset model by \textbf{+5.84\%} using only \textbf{5\%} of the data, while our aligned sampling strategy further boosts average performance by \textbf{+4.24\%}.
A Generative Foundation Model for Chest Radiography
Sep 04, 2025Abstract:The scarcity of well-annotated diverse medical images is a major hurdle for developing reliable AI models in healthcare. Substantial technical advances have been made in generative foundation models for natural images. Here we develop `ChexGen', a generative vision-language foundation model that introduces a unified framework for text-, mask-, and bounding box-guided synthesis of chest radiographs. Built upon the latent diffusion transformer architecture, ChexGen was pretrained on the largest curated chest X-ray dataset to date, consisting of 960,000 radiograph-report pairs. ChexGen achieves accurate synthesis of radiographs through expert evaluations and quantitative metrics. We demonstrate the utility of ChexGen for training data augmentation and supervised pretraining, which led to performance improvements across disease classification, detection, and segmentation tasks using a small fraction of training data. Further, our model enables the creation of diverse patient cohorts that enhance model fairness by detecting and mitigating demographic biases. Our study supports the transformative role of generative foundation models in building more accurate, data-efficient, and equitable medical AI systems.
WSI-LLaVA: A Multimodal Large Language Model for Whole Slide Image
Dec 03, 2024



Abstract:Recent advancements in computational pathology have produced patch-level Multi-modal Large Language Models (MLLMs), but these models are limited by their inability to analyze whole slide images (WSIs) comprehensively and their tendency to bypass crucial morphological features that pathologists rely on for diagnosis. To address these challenges, we first introduce WSI-Bench, a large-scale morphology-aware benchmark containing 180k VQA pairs from 9,850 WSIs across 30 cancer types, designed to evaluate MLLMs' understanding of morphological characteristics crucial for accurate diagnosis. Building upon this benchmark, we present WSI-LLaVA, a novel framework for gigapixel WSI understanding that employs a three-stage training approach: WSI-text alignment, feature space alignment, and task-specific instruction tuning. To better assess model performance in pathological contexts, we develop two specialized WSI metrics: WSI-Precision and WSI-Relevance. Experimental results demonstrate that WSI-LLaVA outperforms existing models across all capability dimensions, with a significant improvement in morphological analysis, establishing a clear correlation between morphological understanding and diagnostic accuracy.
Artificial Intelligence-Enhanced Couinaud Segmentation for Precision Liver Cancer Therapy
Nov 05, 2024



Abstract:Precision therapy for liver cancer necessitates accurately delineating liver sub-regions to protect healthy tissue while targeting tumors, which is essential for reducing recurrence and improving survival rates. However, the segmentation of hepatic segments, known as Couinaud segmentation, is challenging due to indistinct sub-region boundaries and the need for extensive annotated datasets. This study introduces LiverFormer, a novel Couinaud segmentation model that effectively integrates global context with low-level local features based on a 3D hybrid CNN-Transformer architecture. Additionally, a registration-based data augmentation strategy is equipped to enhance the segmentation performance with limited labeled data. Evaluated on CT images from 123 patients, LiverFormer demonstrated high accuracy and strong concordance with expert annotations across various metrics, allowing for enhanced treatment planning for surgery and radiation therapy. It has great potential to reduces complications and minimizes potential damages to surrounding tissue, leading to improved outcomes for patients undergoing complex liver cancer treatments.
Domain generalization across tumor types, laboratories, and species -- insights from the 2022 edition of the Mitosis Domain Generalization Challenge
Sep 27, 2023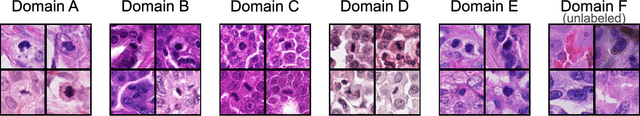
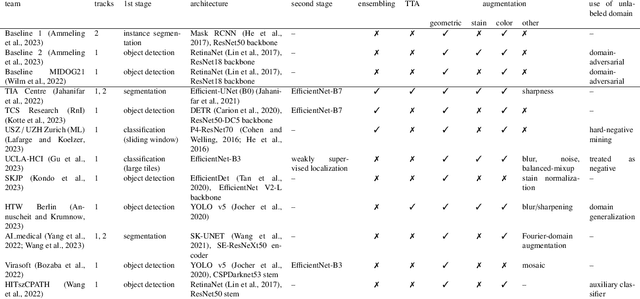
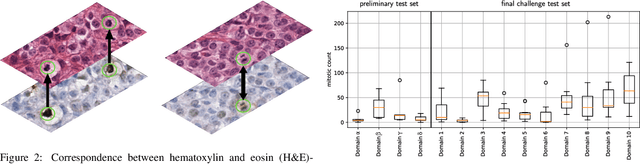
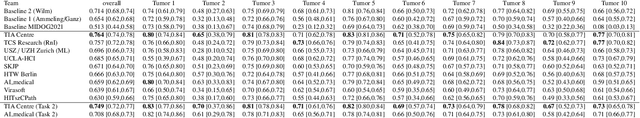
Abstract:Recognition of mitotic figures in histologic tumor specimens is highly relevant to patient outcome assessment. This task is challenging for algorithms and human experts alike, with deterioration of algorithmic performance under shifts in image representations. Considerable covariate shifts occur when assessment is performed on different tumor types, images are acquired using different digitization devices, or specimens are produced in different laboratories. This observation motivated the inception of the 2022 challenge on MItosis Domain Generalization (MIDOG 2022). The challenge provided annotated histologic tumor images from six different domains and evaluated the algorithmic approaches for mitotic figure detection provided by nine challenge participants on ten independent domains. Ground truth for mitotic figure detection was established in two ways: a three-expert consensus and an independent, immunohistochemistry-assisted set of labels. This work represents an overview of the challenge tasks, the algorithmic strategies employed by the participants, and potential factors contributing to their success. With an $F_1$ score of 0.764 for the top-performing team, we summarize that domain generalization across various tumor domains is possible with today's deep learning-based recognition pipelines. When assessed against the immunohistochemistry-assisted reference standard, all methods resulted in reduced recall scores, but with only minor changes in the order of participants in the ranking.
Why is the winner the best?
Mar 30, 2023



Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Federated contrastive learning models for prostate cancer diagnosis and Gleason grading
Feb 17, 2023



Abstract:The application effect of artificial intelligence (AI) in the field of medical imaging is remarkable. Robust AI model training requires large datasets, but data collection faces communication, ethics, and privacy protection constraints. Fortunately, federated learning can solve the above problems by coordinating multiple clients to train the model without sharing the original data. In this study, we design a federated contrastive learning framework (FCL) for large-scale pathology images and the heterogeneity challenges. It enhances the model's generalization ability by maximizing the attention consistency between the local client and server models. To alleviate the privacy leakage problem when transferring parameters and verify the robustness of FCL, we use differential privacy to further protect the model by adding noise. We evaluate the effectiveness of FCL on the cancer diagnosis task and Gleason grading task on 19,635 prostate cancer WSIs from multiple clients. In the diagnosis task, the average AUC of 7 clients is 95% when the categories are relatively balanced, and our FCL achieves 97%. In the Gleason grading task, the average Kappa of 6 clients is 0.74, and the Kappa of FCL reaches 0.84. Furthermore, we also validate the robustness of the model on external datasets(one public dataset and two private datasets). In addition, to better explain the classification effect of the model, we show whether the model focuses on the lesion area by drawing a heatmap. Finally, FCL brings a robust, accurate, low-cost AI training model to biomedical research, effectively protecting medical data privacy.
Artificial intelligence for diagnosing and predicting survival of patients with renal cell carcinoma: Retrospective multi-center study
Jan 12, 2023



Abstract:Background: Clear cell renal cell carcinoma (ccRCC) is the most common renal-related tumor with high heterogeneity. There is still an urgent need for novel diagnostic and prognostic biomarkers for ccRCC. Methods: We proposed a weakly-supervised deep learning strategy using conventional histology of 1752 whole slide images from multiple centers. Our study was demonstrated through internal cross-validation and external validations for the deep learning-based models. Results: Automatic diagnosis for ccRCC through intelligent subtyping of renal cell carcinoma was proved in this study. Our graderisk achieved aera the curve (AUC) of 0.840 (95% confidence interval: 0.805-0.871) in the TCGA cohort, 0.840 (0.805-0.871) in the General cohort, and 0.840 (0.805-0.871) in the CPTAC cohort for the recognition of high-grade tumor. The OSrisk for the prediction of 5-year survival status achieved AUC of 0.784 (0.746-0.819) in the TCGA cohort, which was further verified in the independent General cohort and the CPTAC cohort, with AUC of 0.774 (0.723-0.820) and 0.702 (0.632-0.765), respectively. Cox regression analysis indicated that graderisk, OSrisk, tumor grade, and tumor stage were found to be independent prognostic factors, which were further incorporated into the competing-risk nomogram (CRN). Kaplan-Meier survival analyses further illustrated that our CRN could significantly distinguish patients with high survival risk, with hazard ratio of 5.664 (3.893-8.239, p < 0.0001) in the TCGA cohort, 35.740 (5.889-216.900, p < 0.0001) in the General cohort and 6.107 (1.815 to 20.540, p < 0.0001) in the CPTAC cohort. Comparison analyses conformed that our CRN outperformed current prognosis indicators in the prediction of survival status, with higher concordance index for clinical prognosis.
DeepNoise: Disentanglement of Experimental Noise from Real Biological Signals based on Fluorescent Microscopy Image Classification via Deep Learning
Sep 13, 2022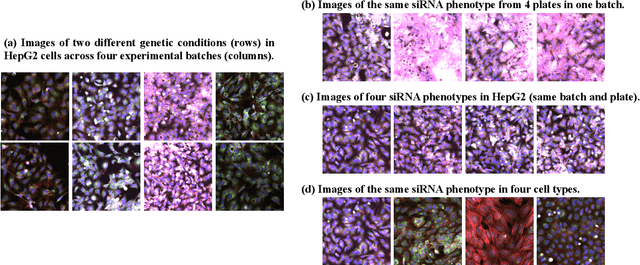
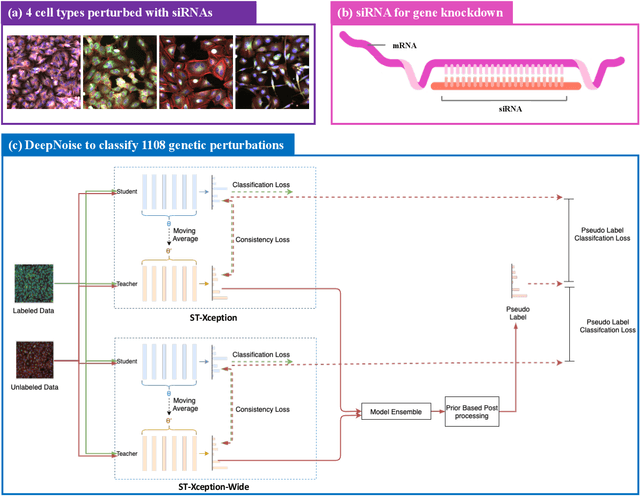
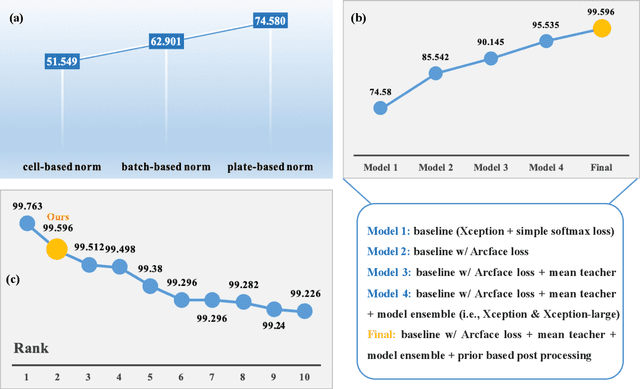
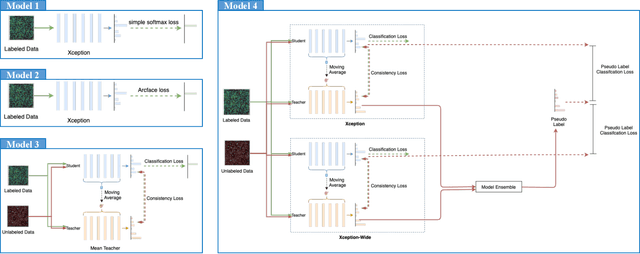
Abstract:The high-content image-based assay is commonly leveraged for identifying the phenotypic impact of genetic perturbations in biology field. However, a persistent issue remains unsolved during experiments: the interferential technical noise caused by systematic errors (e.g., temperature, reagent concentration, and well location) is always mixed up with the real biological signals, leading to misinterpretation of any conclusion drawn. Here, we show a mean teacher based deep learning model (DeepNoise) that can disentangle biological signals from the experimental noise. Specifically, we aim to classify the phenotypic impact of 1,108 different genetic perturbations screened from 125,510 fluorescent microscopy images, which are totally unrecognizable by human eye. We validate our model by participating in the Recursion Cellular Image Classification Challenge, and our proposed method achieves an extremely high classification score (Acc: 99.596%), ranking the 2nd place among 866 participating groups. This promising result indicates the successful separation of biological and technical factors, which might help decrease the cost of treatment development and expedite the drug discovery process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge