Jinxi Xiang
nnMIL: A generalizable multiple instance learning framework for computational pathology
Nov 18, 2025Abstract:Computational pathology holds substantial promise for improving diagnosis and guiding treatment decisions. Recent pathology foundation models enable the extraction of rich patch-level representations from large-scale whole-slide images (WSIs), but current approaches for aggregating these features into slide-level predictions remain constrained by design limitations that hinder generalizability and reliability. Here, we developed nnMIL, a simple yet broadly applicable multiple-instance learning framework that connects patch-level foundation models to robust slide-level clinical inference. nnMIL introduces random sampling at both the patch and feature levels, enabling large-batch optimization, task-aware sampling strategies, and efficient and scalable training across datasets and model architectures. A lightweight aggregator performs sliding-window inference to generate ensemble slide-level predictions and supports principled uncertainty estimation. Across 40,000 WSIs encompassing 35 clinical tasks and four pathology foundation models, nnMIL consistently outperformed existing MIL methods for disease diagnosis, histologic subtyping, molecular biomarker detection, and pan- cancer prognosis prediction. It further demonstrated strong cross-model generalization, reliable uncertainty quantification, and robust survival stratification in multiple external cohorts. In conclusion, nnMIL offers a practical and generalizable solution for translating pathology foundation models into clinically meaningful predictions, advancing the development and deployment of reliable AI systems in real-world settings.
Image Super-resolution Via Latent Diffusion: A Sampling-space Mixture Of Experts And Frequency-augmented Decoder Approach
Oct 20, 2023



Abstract:The recent use of diffusion prior, enhanced by pre-trained text-image models, has markedly elevated the performance of image super-resolution (SR). To alleviate the huge computational cost required by pixel-based diffusion SR, latent-based methods utilize a feature encoder to transform the image and then implement the SR image generation in a compact latent space. Nevertheless, there are two major issues that limit the performance of latent-based diffusion. First, the compression of latent space usually causes reconstruction distortion. Second, huge computational cost constrains the parameter scale of the diffusion model. To counteract these issues, we first propose a frequency compensation module that enhances the frequency components from latent space to pixel space. The reconstruction distortion (especially for high-frequency information) can be significantly decreased. Then, we propose to use Sample-Space Mixture of Experts (SS-MoE) to achieve more powerful latent-based SR, which steadily improves the capacity of the model without a significant increase in inference costs. These carefully crafted designs contribute to performance improvements in largely explored 4x blind super-resolution benchmarks and extend to large magnification factors, i.e., 8x image SR benchmarks. The code is available at https://github.com/amandaluof/moe_sr.
Effortless Cross-Platform Video Codec: A Codebook-Based Method
Oct 16, 2023Abstract:Under certain circumstances, advanced neural video codecs can surpass the most complex traditional codecs in their rate-distortion (RD) performance. One of the main reasons for the high performance of existing neural video codecs is the use of the entropy model, which can provide more accurate probability distribution estimations for compressing the latents. This also implies the rigorous requirement that entropy models running on different platforms should use consistent distribution estimations. However, in cross-platform scenarios, entropy models running on different platforms usually yield inconsistent probability distribution estimations due to floating point computation errors that are platform-dependent, which can cause the decoding side to fail in correctly decoding the compressed bitstream sent by the encoding side. In this paper, we propose a cross-platform video compression framework based on codebooks, which avoids autoregressive entropy modeling and achieves video compression by transmitting the index sequence of the codebooks. Moreover, instead of using optical flow for context alignment, we propose to use the conditional cross-attention module to obtain the context between frames. Due to the absence of autoregressive modeling and optical flow alignment, we can design an extremely minimalist framework that can greatly benefit computational efficiency. Importantly, our framework no longer contains any distribution estimation modules for entropy modeling, and thus computations across platforms are not necessarily consistent. Experimental results show that our method can outperform the traditional H.265 (medium) even without any entropy constraints, while achieving the cross-platform property intrinsically.
Towards Real-Time Neural Video Codec for Cross-Platform Application Using Calibration Information
Sep 20, 2023



Abstract:The state-of-the-art neural video codecs have outperformed the most sophisticated traditional codecs in terms of RD performance in certain cases. However, utilizing them for practical applications is still challenging for two major reasons. 1) Cross-platform computational errors resulting from floating point operations can lead to inaccurate decoding of the bitstream. 2) The high computational complexity of the encoding and decoding process poses a challenge in achieving real-time performance. In this paper, we propose a real-time cross-platform neural video codec, which is capable of efficiently decoding of 720P video bitstream from other encoding platforms on a consumer-grade GPU. First, to solve the problem of inconsistency of codec caused by the uncertainty of floating point calculations across platforms, we design a calibration transmitting system to guarantee the consistent quantization of entropy parameters between the encoding and decoding stages. The parameters that may have transboundary quantization between encoding and decoding are identified in the encoding stage, and their coordinates will be delivered by auxiliary transmitted bitstream. By doing so, these inconsistent parameters can be processed properly in the decoding stage. Furthermore, to reduce the bitrate of the auxiliary bitstream, we rectify the distribution of entropy parameters using a piecewise Gaussian constraint. Second, to match the computational limitations on the decoding side for real-time video codec, we design a lightweight model. A series of efficiency techniques enable our model to achieve 25 FPS decoding speed on NVIDIA RTX 2080 GPU. Experimental results demonstrate that our model can achieve real-time decoding of 720P videos while encoding on another platform. Furthermore, the real-time model brings up to a maximum of 24.2\% BD-rate improvement from the perspective of PSNR with the anchor H.265.
Dynamic Low-Rank Instance Adaptation for Universal Neural Image Compression
Aug 15, 2023



Abstract:The latest advancements in neural image compression show great potential in surpassing the rate-distortion performance of conventional standard codecs. Nevertheless, there exists an indelible domain gap between the datasets utilized for training (i.e., natural images) and those utilized for inference (e.g., artistic images). Our proposal involves a low-rank adaptation approach aimed at addressing the rate-distortion drop observed in out-of-domain datasets. Specifically, we perform low-rank matrix decomposition to update certain adaptation parameters of the client's decoder. These updated parameters, along with image latents, are encoded into a bitstream and transmitted to the decoder in practical scenarios. Due to the low-rank constraint imposed on the adaptation parameters, the resulting bit rate overhead is small. Furthermore, the bit rate allocation of low-rank adaptation is \emph{non-trivial}, considering the diverse inputs require varying adaptation bitstreams. We thus introduce a dynamic gating network on top of the low-rank adaptation method, in order to decide which decoder layer should employ adaptation. The dynamic adaptation network is optimized end-to-end using rate-distortion loss. Our proposed method exhibits universality across diverse image datasets. Extensive results demonstrate that this paradigm significantly mitigates the domain gap, surpassing non-adaptive methods with an average BD-rate improvement of approximately $19\%$ across out-of-domain images. Furthermore, it outperforms the most advanced instance adaptive methods by roughly $5\%$ BD-rate. Ablation studies confirm our method's ability to universally enhance various image compression architectures.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Federated contrastive learning models for prostate cancer diagnosis and Gleason grading
Feb 17, 2023



Abstract:The application effect of artificial intelligence (AI) in the field of medical imaging is remarkable. Robust AI model training requires large datasets, but data collection faces communication, ethics, and privacy protection constraints. Fortunately, federated learning can solve the above problems by coordinating multiple clients to train the model without sharing the original data. In this study, we design a federated contrastive learning framework (FCL) for large-scale pathology images and the heterogeneity challenges. It enhances the model's generalization ability by maximizing the attention consistency between the local client and server models. To alleviate the privacy leakage problem when transferring parameters and verify the robustness of FCL, we use differential privacy to further protect the model by adding noise. We evaluate the effectiveness of FCL on the cancer diagnosis task and Gleason grading task on 19,635 prostate cancer WSIs from multiple clients. In the diagnosis task, the average AUC of 7 clients is 95% when the categories are relatively balanced, and our FCL achieves 97%. In the Gleason grading task, the average Kappa of 6 clients is 0.74, and the Kappa of FCL reaches 0.84. Furthermore, we also validate the robustness of the model on external datasets(one public dataset and two private datasets). In addition, to better explain the classification effect of the model, we show whether the model focuses on the lesion area by drawing a heatmap. Finally, FCL brings a robust, accurate, low-cost AI training model to biomedical research, effectively protecting medical data privacy.
Artificial intelligence for diagnosing and predicting survival of patients with renal cell carcinoma: Retrospective multi-center study
Jan 12, 2023



Abstract:Background: Clear cell renal cell carcinoma (ccRCC) is the most common renal-related tumor with high heterogeneity. There is still an urgent need for novel diagnostic and prognostic biomarkers for ccRCC. Methods: We proposed a weakly-supervised deep learning strategy using conventional histology of 1752 whole slide images from multiple centers. Our study was demonstrated through internal cross-validation and external validations for the deep learning-based models. Results: Automatic diagnosis for ccRCC through intelligent subtyping of renal cell carcinoma was proved in this study. Our graderisk achieved aera the curve (AUC) of 0.840 (95% confidence interval: 0.805-0.871) in the TCGA cohort, 0.840 (0.805-0.871) in the General cohort, and 0.840 (0.805-0.871) in the CPTAC cohort for the recognition of high-grade tumor. The OSrisk for the prediction of 5-year survival status achieved AUC of 0.784 (0.746-0.819) in the TCGA cohort, which was further verified in the independent General cohort and the CPTAC cohort, with AUC of 0.774 (0.723-0.820) and 0.702 (0.632-0.765), respectively. Cox regression analysis indicated that graderisk, OSrisk, tumor grade, and tumor stage were found to be independent prognostic factors, which were further incorporated into the competing-risk nomogram (CRN). Kaplan-Meier survival analyses further illustrated that our CRN could significantly distinguish patients with high survival risk, with hazard ratio of 5.664 (3.893-8.239, p < 0.0001) in the TCGA cohort, 35.740 (5.889-216.900, p < 0.0001) in the General cohort and 6.107 (1.815 to 20.540, p < 0.0001) in the CPTAC cohort. Comparison analyses conformed that our CRN outperformed current prognosis indicators in the prediction of survival status, with higher concordance index for clinical prognosis.
Pan-cancer computational histopathology reveals tumor mutational burden status through weakly-supervised deep learning
Apr 07, 2022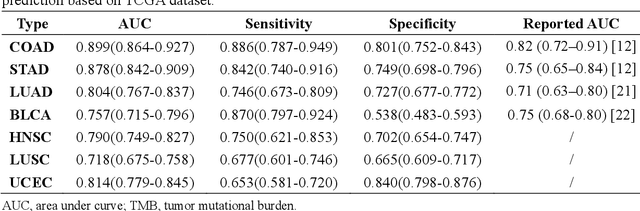
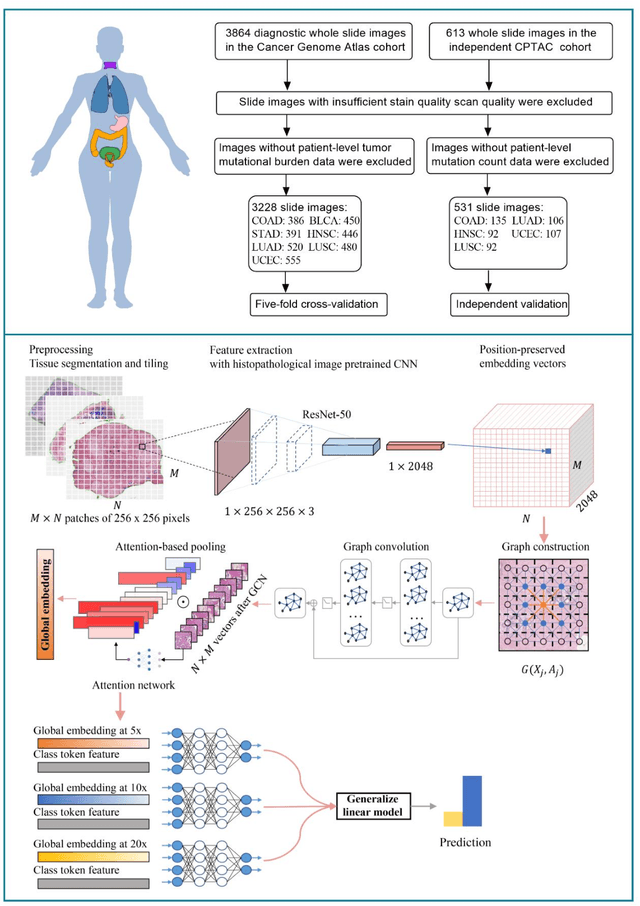
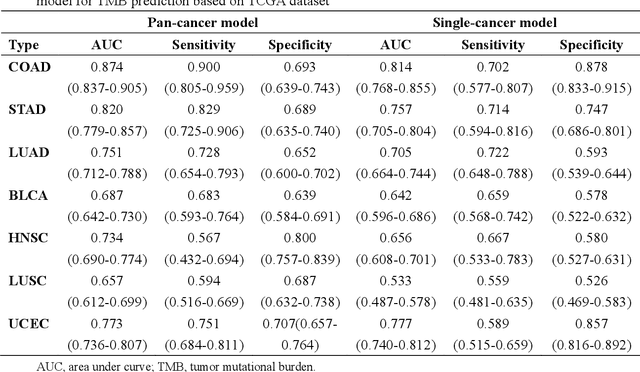
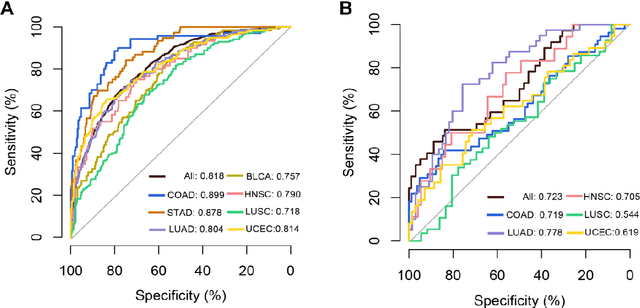
Abstract:Tumor mutational burden (TMB) is a potential genomic biomarker that can help identify patients who will benefit from immunotherapy across a variety of cancers. We included whole slide images (WSIs) of 3228 diagnostic slides from the Cancer Genome Atlas and 531 WSIs from the Clinical Proteomic Tumor Analysis Consortium for the development and verification of a pan-cancer TMB prediction model (PC-TMB). We proposed a multiscale weakly-supervised deep learning framework for predicting TMB of seven types of tumors based only on routinely used hematoxylin-eosin (H&E)-stained WSIs. PC-TMB achieved a mean area under curve (AUC) of 0.818 (0.804-0.831) in the cross-validation cohort, which was superior to the best single-scale model. In comparison with the state-of-the-art TMB prediction model from previous publications, our multiscale model achieved better performance over previously reported models. In addition, the improvements of PC-TMB over the single-tumor models were also confirmed by the ablation tests on 10x magnification. The PC-TMB algorithm also exhibited good generalization on external validation cohort with AUC of 0.732 (0.683-0.761). PC-TMB possessed a comparable survival-risk stratification performance to the TMB measured by whole exome sequencing, but with low cost and being time-efficient for providing a prognostic biomarker of multiple solid tumors. Moreover, spatial heterogeneity of TMB within tumors was also identified through our PC-TMB, which might enable image-based screening for molecular biomarkers with spatial variation and potential exploring for genotype-spatial heterogeneity relationships.
Self-Ensembling Contrastive Learning for Semi-Supervised Medical Image Segmentation
Jun 10, 2021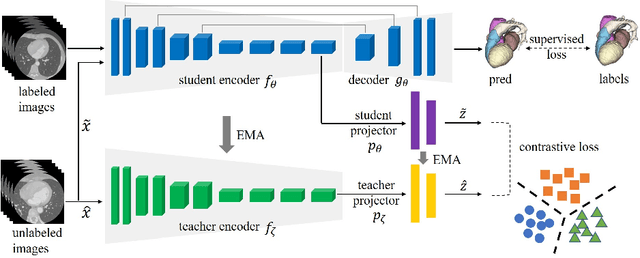
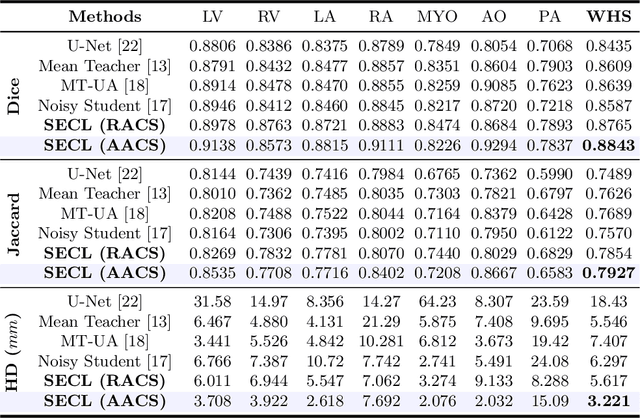
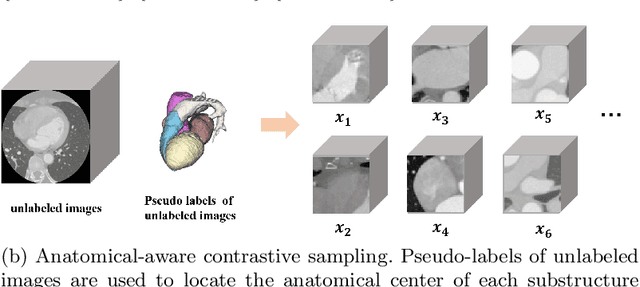
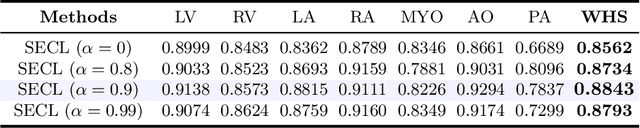
Abstract:Deep learning has demonstrated significant improvements in medical image segmentation using a sufficiently large amount of training data with manual labels. Acquiring well-representative labels requires expert knowledge and exhaustive labors. In this paper, we aim to boost the performance of semi-supervised learning for medical image segmentation with limited labels using a self-ensembling contrastive learning technique. To this end, we propose to train an encoder-decoder network at image-level with small amounts of labeled images, and more importantly, we learn latent representations directly at feature-level by imposing contrastive loss on unlabeled images. This method strengthens intra-class compactness and inter-class separability, so as to get a better pixel classifier. Moreover, we devise a student encoder for online learning and an exponential moving average version of it, called teacher encoder, to improve the performance iteratively in a self-ensembling manner. To construct contrastive samples with unlabeled images, two sampling strategies that exploit structure similarity across medical images and utilize pseudo-labels for construction, termed region-aware and anatomical-aware contrastive sampling, are investigated. We conduct extensive experiments on an MRI and a CT segmentation dataset and demonstrate that in a limited label setting, the proposed method achieves state-of-the-art performance. Moreover, the anatomical-aware strategy that prepares contrastive samples on-the-fly using pseudo-labels realizes better contrastive regularization on feature representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge